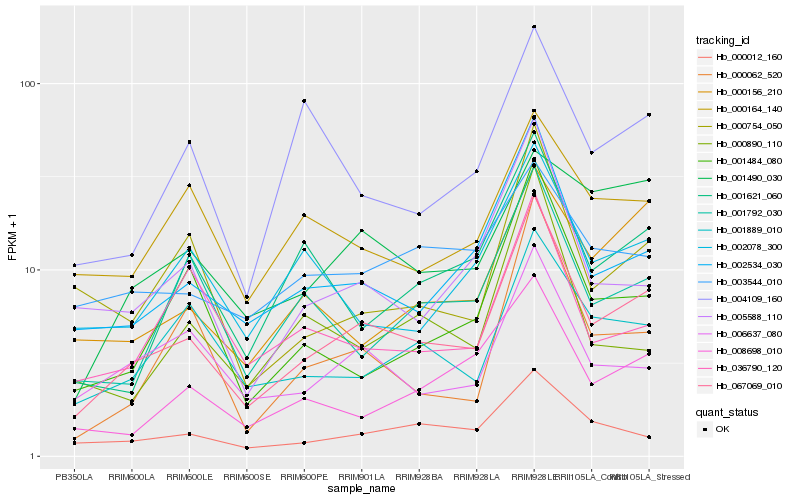
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_067069_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 2 |
Hb_002534_030 |
0.1260491447 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 3 |
Hb_001889_010 |
0.1611776676 |
transcription factor |
TF Family: Orphans |
PREDICTED: protein CHLOROPLAST IMPORT APPARATUS 2 [Jatropha curcas] |
| 4 |
Hb_005588_110 |
0.1618181798 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 5 |
Hb_008698_010 |
0.161847009 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_000012_160 |
0.1631372649 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006637_080 |
0.1644140321 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001484_080 |
0.1681660149 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 9 |
Hb_000062_520 |
0.1687965109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000754_050 |
0.1694938451 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001792_030 |
0.1710065056 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 12 |
Hb_001621_060 |
0.1720247563 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000156_210 |
0.174075756 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 14 |
Hb_002078_300 |
0.1745694969 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000164_140 |
0.1814974682 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 16 |
Hb_036790_120 |
0.1836868185 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001490_030 |
0.1844763568 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004109_160 |
0.1875235495 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003544_010 |
0.1888477826 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 20 |
Hb_000890_110 |
0.1901205759 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |