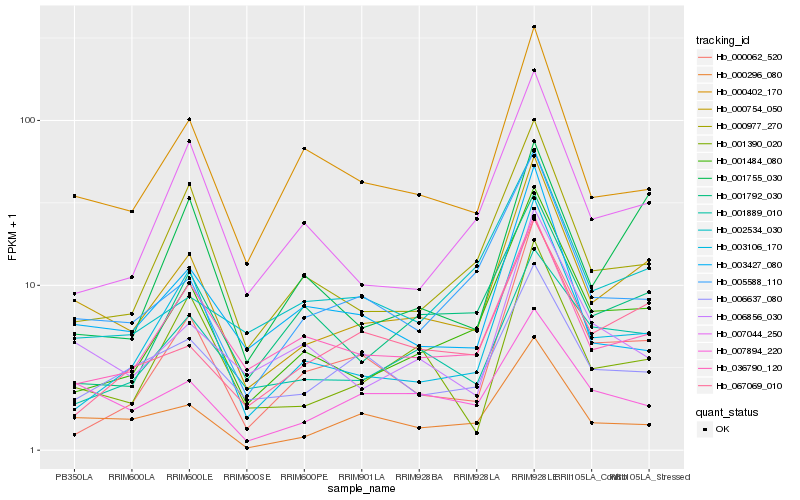
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000754_050 |
0.0 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001484_080 |
0.1310413453 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 3 |
Hb_000296_080 |
0.1391682866 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35130 [Jatropha curcas] |
| 4 |
Hb_005588_110 |
0.1455714176 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 5 |
Hb_000402_170 |
0.1494120734 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 6 |
Hb_036790_120 |
0.1497296322 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000977_270 |
0.1544171997 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003106_170 |
0.1547402518 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 9 |
Hb_006856_030 |
0.1584715393 |
- |
- |
Uridine kinase-like protein 4 [Glycine soja] |
| 10 |
Hb_007044_250 |
0.1606313037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007894_220 |
0.1607414002 |
- |
- |
PREDICTED: probable folate-biopterin transporter 4 [Jatropha curcas] |
| 12 |
Hb_001889_010 |
0.1623682839 |
transcription factor |
TF Family: Orphans |
PREDICTED: protein CHLOROPLAST IMPORT APPARATUS 2 [Jatropha curcas] |
| 13 |
Hb_001390_020 |
0.165746268 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_006637_080 |
0.1669057043 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001792_030 |
0.169432231 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 16 |
Hb_067069_010 |
0.1694938451 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 17 |
Hb_002534_030 |
0.1703231837 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 18 |
Hb_001755_030 |
0.1713799372 |
- |
- |
PREDICTED: D-glycerate 3-kinase, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_000062_520 |
0.1716372971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003427_080 |
0.1731003686 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |