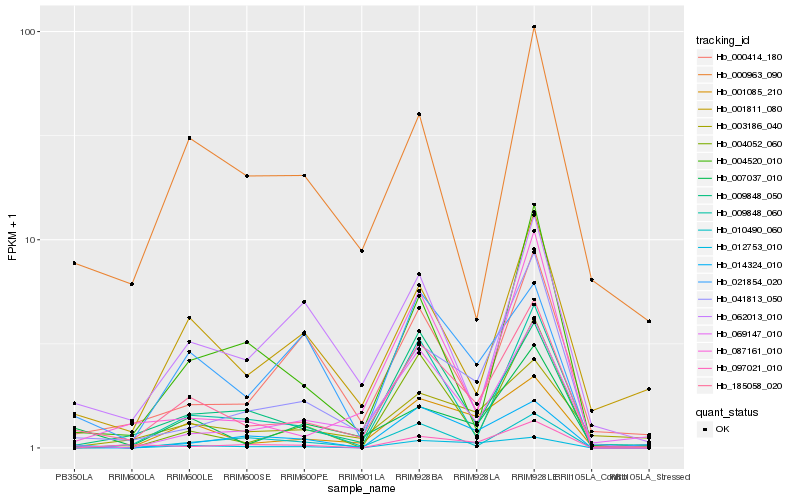
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185058_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 2 |
Hb_009848_050 |
0.1691951827 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK13 [Elaeis guineensis] |
| 3 |
Hb_001085_210 |
0.1732477735 |
- |
- |
PREDICTED: myosin-binding protein 3 [Jatropha curcas] |
| 4 |
Hb_007037_010 |
0.1774204171 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 5 |
Hb_041813_050 |
0.1830610122 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 6 |
Hb_009848_060 |
0.1950263485 |
- |
- |
unknown [Glycine max] |
| 7 |
Hb_069147_010 |
0.1996448189 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At4g27220 isoform X1 [Populus euphratica] |
| 8 |
Hb_010490_060 |
0.2050920539 |
- |
- |
PREDICTED: uncharacterized protein LOC105767021 [Gossypium raimondii] |
| 9 |
Hb_001811_080 |
0.2051755835 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 10 |
Hb_003186_040 |
0.2097248974 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |
| 11 |
Hb_004520_010 |
0.2109666989 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 12 |
Hb_087161_010 |
0.2119029121 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_021854_020 |
0.2122529268 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 14 |
Hb_012753_010 |
0.216415298 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_004052_060 |
0.2173945515 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_062013_010 |
0.2191650154 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_000414_180 |
0.2223488682 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 18 |
Hb_097021_010 |
0.2336691717 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 19 |
Hb_000963_090 |
0.2352935873 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_014324_010 |
0.2379378833 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |