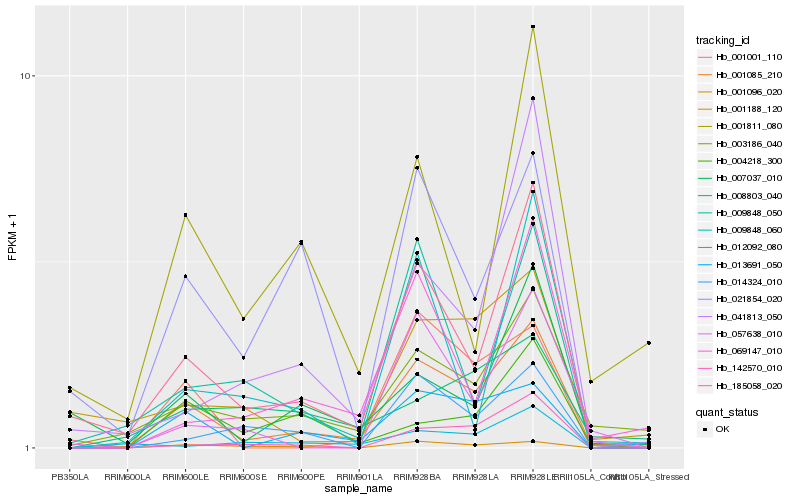
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_210 |
0.0 |
- |
- |
PREDICTED: myosin-binding protein 3 [Jatropha curcas] |
| 2 |
Hb_185058_020 |
0.1732477735 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 3 |
Hb_013691_050 |
0.1750740131 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 4 |
Hb_003186_040 |
0.1775350988 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |
| 5 |
Hb_001096_020 |
0.2142548696 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Populus euphratica] |
| 6 |
Hb_069147_010 |
0.2247337245 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At4g27220 isoform X1 [Populus euphratica] |
| 7 |
Hb_142570_010 |
0.2410844873 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_001001_110 |
0.241526179 |
- |
- |
- |
| 9 |
Hb_014324_010 |
0.2450370452 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 10 |
Hb_007037_010 |
0.2473123076 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 11 |
Hb_001188_120 |
0.250374684 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 12 |
Hb_021854_020 |
0.2508148716 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 13 |
Hb_009848_050 |
0.2522814633 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK13 [Elaeis guineensis] |
| 14 |
Hb_001811_080 |
0.2523727174 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 15 |
Hb_041813_050 |
0.2538860937 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 16 |
Hb_057638_010 |
0.2562000608 |
- |
- |
PREDICTED: uncharacterized protein LOC105637008 [Jatropha curcas] |
| 17 |
Hb_009848_060 |
0.2575728179 |
- |
- |
unknown [Glycine max] |
| 18 |
Hb_004218_300 |
0.2596518848 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 19 |
Hb_012092_080 |
0.2598461191 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 20 |
Hb_008803_040 |
0.2612197372 |
- |
- |
PREDICTED: uncharacterized protein LOC105157106 [Sesamum indicum] |