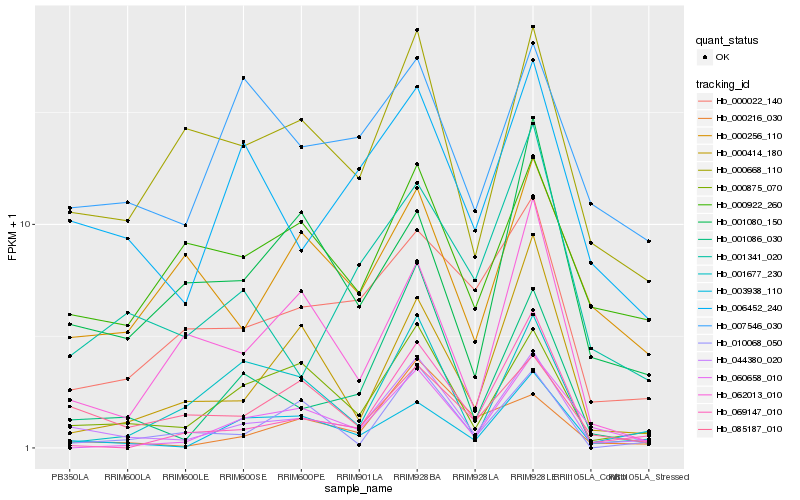
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044380_020 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_001086_030 |
0.145574075 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 3 |
Hb_000875_070 |
0.1541121905 |
- |
- |
- |
| 4 |
Hb_000668_110 |
0.1829501127 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 5 |
Hb_006452_240 |
0.1864695213 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 15a isoform X1 [Jatropha curcas] |
| 6 |
Hb_003938_110 |
0.2061701185 |
- |
- |
polyprotein [Ananas comosus] |
| 7 |
Hb_000414_180 |
0.2072026804 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 8 |
Hb_069147_010 |
0.2074506708 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At4g27220 isoform X1 [Populus euphratica] |
| 9 |
Hb_010068_050 |
0.2082570351 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 10 |
Hb_000022_140 |
0.2089779918 |
- |
- |
hypothetical protein MIMGU_mgv1a009920mg [Erythranthe guttata] |
| 11 |
Hb_001341_020 |
0.2090047397 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 12 |
Hb_085187_010 |
0.2124375733 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 13 |
Hb_060658_010 |
0.2153530132 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 14 |
Hb_062013_010 |
0.2156488947 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 15 |
Hb_000216_030 |
0.2175334774 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 16 |
Hb_001080_150 |
0.2210412855 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 17 |
Hb_001677_230 |
0.2221263172 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000922_260 |
0.2239936867 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 19 |
Hb_000256_110 |
0.2330707364 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007546_030 |
0.2344304028 |
- |
- |
transcription cofactor, putative [Ricinus communis] |