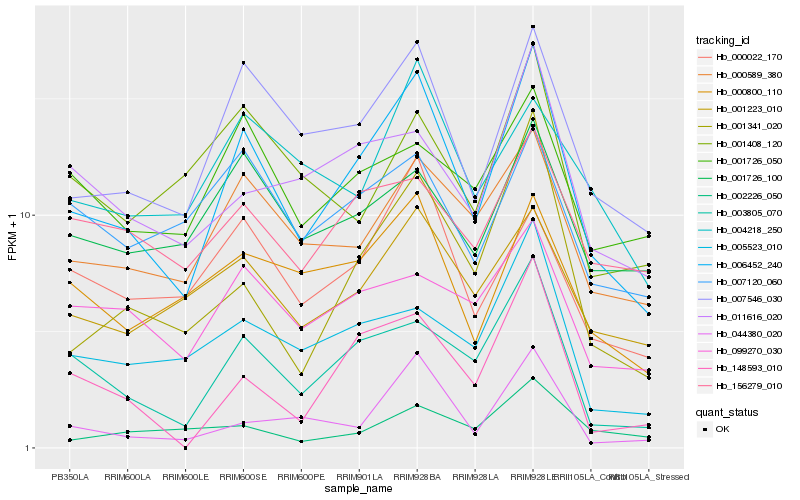
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006452_240 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 15a isoform X1 [Jatropha curcas] |
| 2 |
Hb_007546_030 |
0.1210880038 |
- |
- |
transcription cofactor, putative [Ricinus communis] |
| 3 |
Hb_003805_070 |
0.1300103827 |
- |
- |
PREDICTED: sacsin [Jatropha curcas] |
| 4 |
Hb_000589_380 |
0.1490759338 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 5 |
Hb_001341_020 |
0.1574502689 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 6 |
Hb_001223_010 |
0.1615288115 |
- |
- |
PREDICTED: FGGY carbohydrate kinase domain-containing protein isoform X2 [Jatropha curcas] |
| 7 |
Hb_000800_110 |
0.1621572167 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_005523_010 |
0.1650730216 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 9 |
Hb_000022_170 |
0.1685505683 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 10 |
Hb_011616_020 |
0.1728970483 |
- |
- |
hypothetical protein CICLE_v10004968mg [Citrus clementina] |
| 11 |
Hb_001726_100 |
0.1759660203 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 12 |
Hb_099270_030 |
0.1761916386 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 13 |
Hb_148593_010 |
0.1768114685 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR-like [Jatropha curcas] |
| 14 |
Hb_156279_010 |
0.1823842678 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007120_060 |
0.1829677 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 16 |
Hb_004218_250 |
0.1856740921 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 17 |
Hb_001726_050 |
0.186191239 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 18 |
Hb_001408_120 |
0.1863939608 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_044380_020 |
0.1864695213 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 20 |
Hb_002226_050 |
0.1883296432 |
- |
- |
PREDICTED: uncharacterized protein LOC105641366 isoform X1 [Jatropha curcas] |