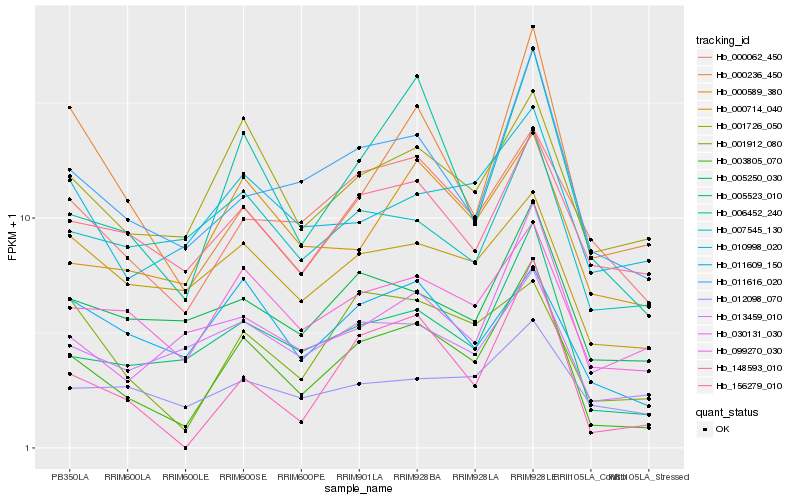
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003805_070 |
0.0 |
- |
- |
PREDICTED: sacsin [Jatropha curcas] |
| 2 |
Hb_006452_240 |
0.1300103827 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 15a isoform X1 [Jatropha curcas] |
| 3 |
Hb_005523_010 |
0.131666711 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 4 |
Hb_148593_010 |
0.1453236021 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR-like [Jatropha curcas] |
| 5 |
Hb_011616_020 |
0.1480240042 |
- |
- |
hypothetical protein CICLE_v10004968mg [Citrus clementina] |
| 6 |
Hb_099270_030 |
0.1549811425 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001912_080 |
0.1729217086 |
- |
- |
vacuolar protein sorting-associated protein, putative [Ricinus communis] |
| 8 |
Hb_001726_050 |
0.1747733618 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_000589_380 |
0.1766150277 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 10 |
Hb_005250_030 |
0.1787641614 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 11 |
Hb_000236_450 |
0.1819202837 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 12 |
Hb_000062_450 |
0.1819968489 |
- |
- |
hypothetical protein MTR_081s0009, partial [Medicago truncatula] |
| 13 |
Hb_013459_010 |
0.1866812424 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 14 |
Hb_156279_010 |
0.1870716506 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |
| 15 |
Hb_030131_030 |
0.1910124695 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g42310, mitochondrial [Jatropha curcas] |
| 16 |
Hb_011609_150 |
0.1912992805 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |
| 17 |
Hb_000714_040 |
0.1916507132 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 18 |
Hb_010998_020 |
0.1928231797 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 19 |
Hb_007545_130 |
0.1928688193 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_012098_070 |
0.1960060923 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |