| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
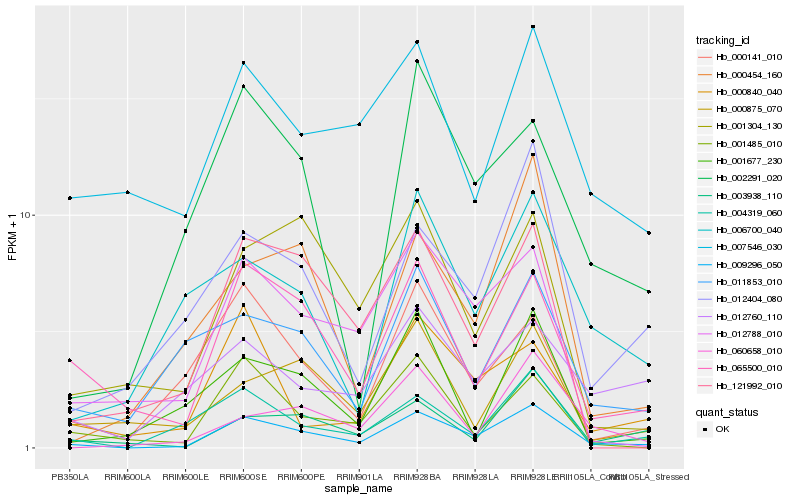
Hb_009296_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 2 |
Hb_065500_010 |
0.1815742799 |
- |
- |
- |
| 3 |
Hb_004319_060 |
0.1909091474 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 4 |
Hb_003938_110 |
0.1999112846 |
- |
- |
polyprotein [Ananas comosus] |
| 5 |
Hb_012404_080 |
0.2033220773 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 6 |
Hb_012788_010 |
0.2040675479 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_121992_010 |
0.2114480369 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 8 |
Hb_000141_010 |
0.2122295678 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 9 |
Hb_001485_010 |
0.2148477997 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 10 |
Hb_000454_160 |
0.2158581325 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001304_130 |
0.219081968 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like transcription factor PAT1 [Jatropha curcas] |
| 12 |
Hb_002291_020 |
0.2195359881 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_060658_010 |
0.22008041 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 14 |
Hb_011853_010 |
0.2202371197 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 15 |
Hb_000840_040 |
0.223283558 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 16 |
Hb_000875_070 |
0.2237412989 |
- |
- |
- |
| 17 |
Hb_006700_040 |
0.2281631474 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 18 |
Hb_007546_030 |
0.2287512308 |
- |
- |
transcription cofactor, putative [Ricinus communis] |
| 19 |
Hb_001677_230 |
0.2301490595 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012760_110 |
0.2308695275 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |