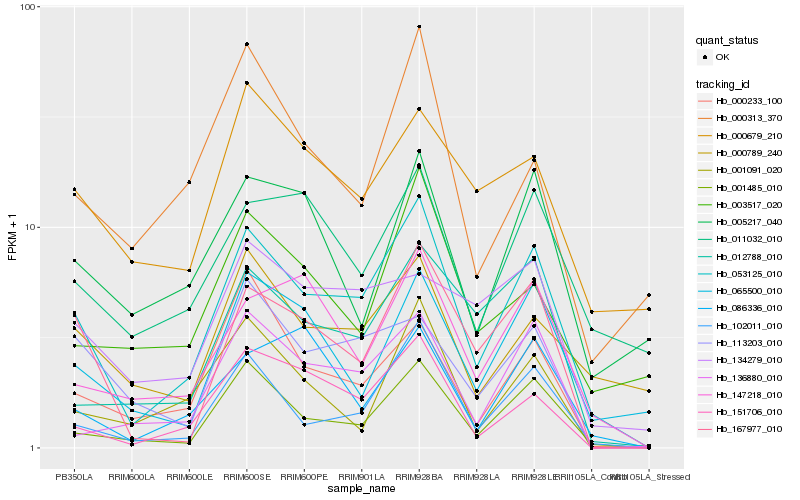
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_053125_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104881508 [Vitis vinifera] |
| 2 |
Hb_001485_010 |
0.142317811 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 3 |
Hb_102011_010 |
0.1440423916 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_167977_010 |
0.1628311922 |
- |
- |
hypothetical protein POPTR_0185s002102g, partial [Populus trichocarpa] |
| 5 |
Hb_151706_010 |
0.1669437132 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 6 |
Hb_147218_010 |
0.1798241485 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_065500_010 |
0.1846573863 |
- |
- |
- |
| 8 |
Hb_136880_010 |
0.1850942803 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_012788_010 |
0.1892880062 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_011032_010 |
0.1955929131 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 11 |
Hb_113203_010 |
0.1959483596 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 12 |
Hb_000233_100 |
0.1964802285 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 13 |
Hb_000789_240 |
0.1983453058 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 14 |
Hb_086336_010 |
0.1986706628 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 15 |
Hb_000679_210 |
0.204199357 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 16 |
Hb_000313_370 |
0.2042219096 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_003517_020 |
0.2051011471 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 18 |
Hb_001091_020 |
0.206696489 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_005217_040 |
0.2070872924 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 20 |
Hb_134279_010 |
0.2085417121 |
- |
- |
PREDICTED: uncharacterized protein LOC105644390 [Jatropha curcas] |