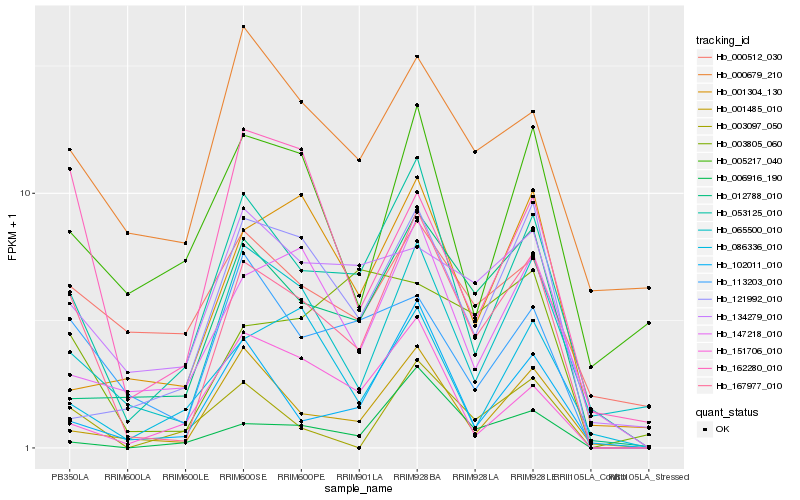
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_167977_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0185s002102g, partial [Populus trichocarpa] |
| 2 |
Hb_053125_010 |
0.1628311922 |
- |
- |
PREDICTED: uncharacterized protein LOC104881508 [Vitis vinifera] |
| 3 |
Hb_065500_010 |
0.2074746993 |
- |
- |
- |
| 4 |
Hb_012788_010 |
0.2141163014 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 5 |
Hb_001485_010 |
0.2167299818 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 6 |
Hb_113203_010 |
0.2189990467 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_102011_010 |
0.2239939218 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_147218_010 |
0.2278425306 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_006916_190 |
0.2290065069 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_003097_050 |
0.2361553472 |
- |
- |
hypothetical protein POPTR_0007s07150g [Populus trichocarpa] |
| 11 |
Hb_134279_010 |
0.2401961853 |
- |
- |
PREDICTED: uncharacterized protein LOC105644390 [Jatropha curcas] |
| 12 |
Hb_121992_010 |
0.2427255883 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 13 |
Hb_001304_130 |
0.2462400182 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like transcription factor PAT1 [Jatropha curcas] |
| 14 |
Hb_162280_010 |
0.2463374884 |
- |
- |
Phosphoprotein phosphatase isoform 2, partial [Theobroma cacao] |
| 15 |
Hb_003805_060 |
0.2469205873 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000679_210 |
0.24859126 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 17 |
Hb_086336_010 |
0.2500492866 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 18 |
Hb_151706_010 |
0.2510392373 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 19 |
Hb_005217_040 |
0.255359141 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000512_030 |
0.2558904361 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |