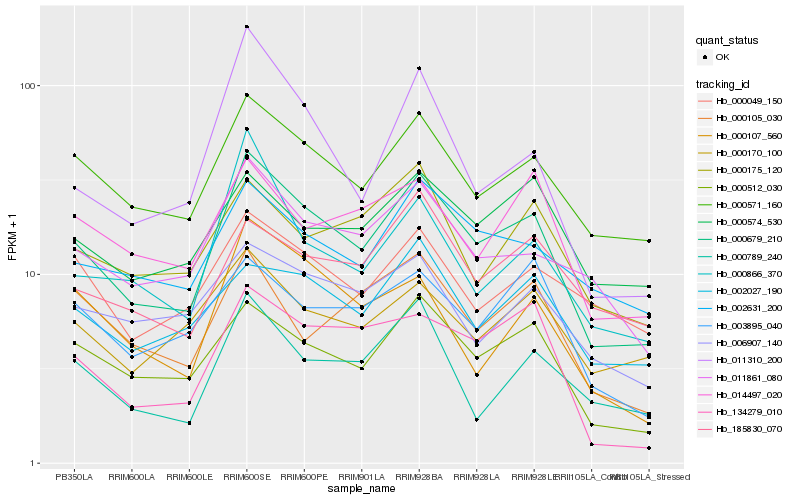
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_210 |
0.0 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 2 |
Hb_000571_160 |
0.1013250442 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 3 |
Hb_000512_030 |
0.10505812 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 4 |
Hb_011861_080 |
0.1187030516 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 5 |
Hb_002631_200 |
0.1294874204 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 6 |
Hb_134279_010 |
0.1299780251 |
- |
- |
PREDICTED: uncharacterized protein LOC105644390 [Jatropha curcas] |
| 7 |
Hb_000789_240 |
0.136429084 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 8 |
Hb_002027_190 |
0.1366632341 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 9 |
Hb_014497_020 |
0.1399563407 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 10 |
Hb_000107_560 |
0.1404436527 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 11 |
Hb_003895_040 |
0.1409230553 |
- |
- |
hypothetical protein CISIN_1g0006512mg, partial [Citrus sinensis] |
| 12 |
Hb_185830_070 |
0.1411812351 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000866_370 |
0.1412519445 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 14 |
Hb_000170_100 |
0.1415255178 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 15 |
Hb_006907_140 |
0.141703709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011310_200 |
0.1420168867 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 17 |
Hb_000049_150 |
0.142596147 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000574_530 |
0.14263166 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 19 |
Hb_000175_120 |
0.1432792045 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 20 |
Hb_000105_030 |
0.1435480352 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |