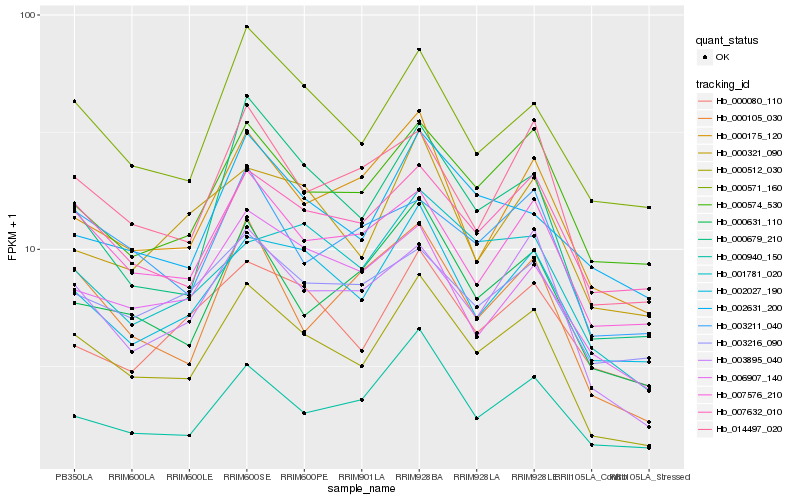
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000512_030 |
0.0 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 2 |
Hb_003895_040 |
0.0906992506 |
- |
- |
hypothetical protein CISIN_1g0006512mg, partial [Citrus sinensis] |
| 3 |
Hb_002027_190 |
0.101424077 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 4 |
Hb_000679_210 |
0.10505812 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 5 |
Hb_000571_160 |
0.1067755411 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 6 |
Hb_014497_020 |
0.1069818807 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 7 |
Hb_000631_110 |
0.1085648107 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 8 |
Hb_007576_210 |
0.111384873 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 9 |
Hb_000105_030 |
0.1117772194 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 10 |
Hb_000175_120 |
0.1138933551 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 11 |
Hb_000574_530 |
0.1140521166 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 12 |
Hb_007632_010 |
0.116269552 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000940_150 |
0.1170061979 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 14 |
Hb_006907_140 |
0.1204881159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000321_090 |
0.1206695107 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 16 |
Hb_001781_020 |
0.1216101149 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 17 |
Hb_003211_040 |
0.1250361905 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002631_200 |
0.1271758649 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003216_090 |
0.1275944375 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000080_110 |
0.128438369 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |