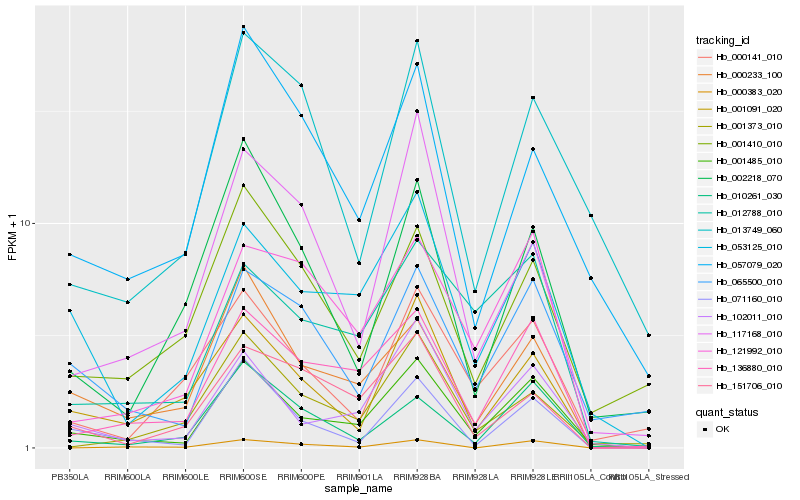
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001485_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 2 |
Hb_102011_010 |
0.1091481271 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_053125_010 |
0.142317811 |
- |
- |
PREDICTED: uncharacterized protein LOC104881508 [Vitis vinifera] |
| 4 |
Hb_136880_010 |
0.1467228034 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 5 |
Hb_071160_010 |
0.1624481496 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 6 |
Hb_000233_100 |
0.166330597 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_065500_010 |
0.1727140603 |
- |
- |
- |
| 8 |
Hb_012788_010 |
0.1795102729 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_001091_020 |
0.1847218774 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 10 |
Hb_010261_030 |
0.1853072095 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 11 |
Hb_121992_010 |
0.1934768757 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 12 |
Hb_001373_010 |
0.1966991472 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 13 |
Hb_000141_010 |
0.1972516823 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 14 |
Hb_057079_020 |
0.1975021546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002218_070 |
0.2023687693 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_013749_060 |
0.2032482385 |
- |
- |
PREDICTED: uncharacterized protein LOC105647762 [Jatropha curcas] |
| 17 |
Hb_001410_010 |
0.2048359848 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 18 |
Hb_117168_010 |
0.2093514625 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 19 |
Hb_151706_010 |
0.2105676942 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 20 |
Hb_000383_020 |
0.2142161146 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |