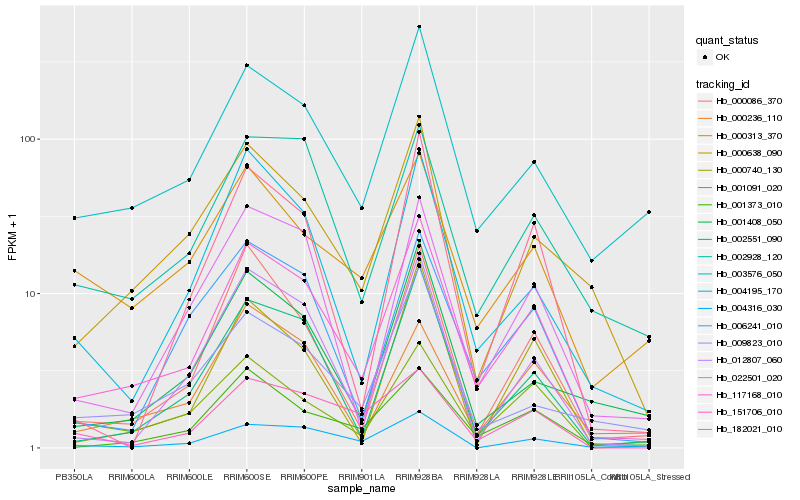
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_117168_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 2 |
Hb_002551_090 |
0.1284340078 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |
| 3 |
Hb_001373_010 |
0.1355790189 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 4 |
Hb_003576_050 |
0.1360517835 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 5 |
Hb_001091_020 |
0.1415304124 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_000086_370 |
0.1419898233 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 7 |
Hb_006241_010 |
0.1450643111 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 8 |
Hb_012807_060 |
0.1453642799 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_022501_020 |
0.145424152 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 10 |
Hb_000638_090 |
0.1478060451 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_004195_170 |
0.1491255992 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_002928_120 |
0.1538157147 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 13 |
Hb_182021_010 |
0.1561731004 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 14 |
Hb_004316_030 |
0.1563386362 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 15 |
Hb_001408_050 |
0.1619505672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000740_130 |
0.162825081 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 17 |
Hb_000236_110 |
0.1639772359 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |
| 18 |
Hb_009823_010 |
0.1666464304 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 19 |
Hb_151706_010 |
0.1676476568 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 20 |
Hb_000313_370 |
0.1679955505 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |