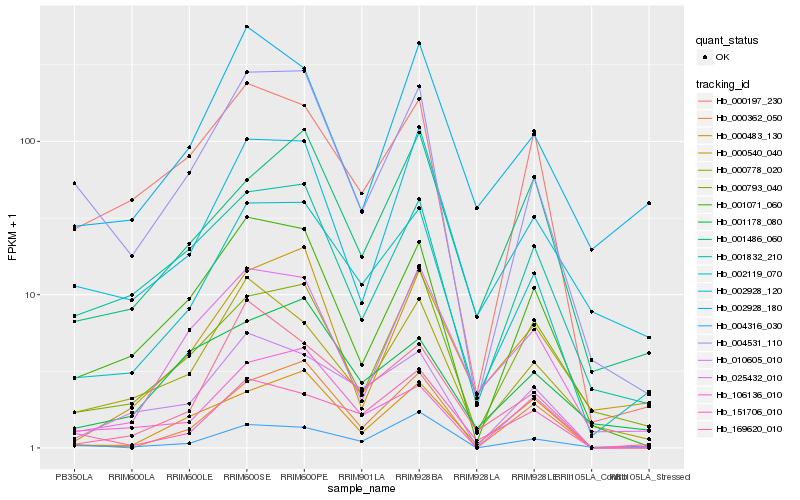
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002119_070 |
0.0 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_010605_010 |
0.134036991 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 3 |
Hb_004531_110 |
0.1489832448 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 4 |
Hb_000362_050 |
0.151491009 |
transcription factor |
TF Family: NOZZLE |
PREDICTED: uncharacterized protein LOC105635737 [Jatropha curcas] |
| 5 |
Hb_002928_120 |
0.1516145631 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 6 |
Hb_001071_060 |
0.1520665681 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 7 |
Hb_151706_010 |
0.1542329379 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 8 |
Hb_004316_030 |
0.1548407855 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 9 |
Hb_025432_010 |
0.1563408682 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 10 |
Hb_000793_040 |
0.1568255854 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 11 |
Hb_001832_210 |
0.1593187621 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 12 |
Hb_169620_010 |
0.1599860302 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 13 |
Hb_000778_020 |
0.16017008 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 14 |
Hb_001486_060 |
0.1605638883 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 15 |
Hb_106136_010 |
0.1614589413 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 16 |
Hb_000197_230 |
0.1631361707 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 17 |
Hb_000483_130 |
0.1672610227 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001178_080 |
0.1691662993 |
- |
- |
PREDICTED: protein kinase PVPK-1 [Jatropha curcas] |
| 19 |
Hb_000540_040 |
0.1721130931 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 20 |
Hb_002928_180 |
0.1724615891 |
- |
- |
catalase CAT1 [Manihot esculenta] |