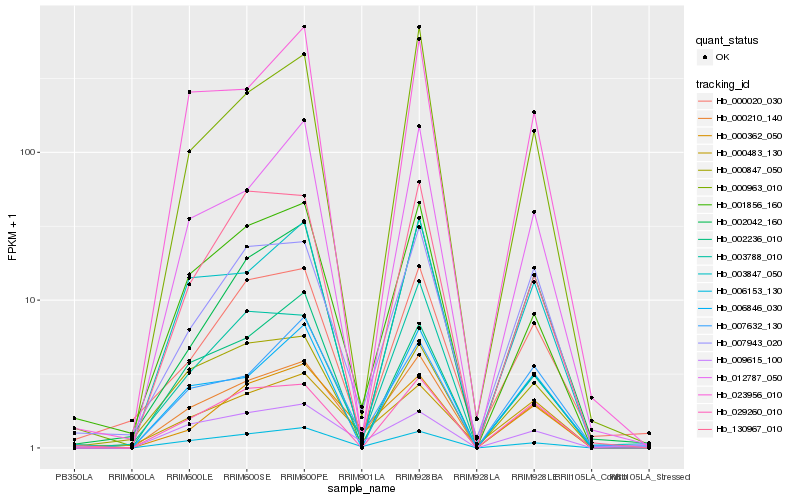
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000210_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 2 |
Hb_006153_130 |
0.0805298026 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_001856_160 |
0.1145513973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003847_050 |
0.125160253 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 5 |
Hb_003788_010 |
0.1278844248 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 6 |
Hb_007943_020 |
0.129424338 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_029260_010 |
0.1318000583 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 8 |
Hb_000963_010 |
0.1328959267 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 9 |
Hb_012787_050 |
0.1350616688 |
- |
- |
PREDICTED: glutathione S-transferase U18-like [Jatropha curcas] |
| 10 |
Hb_000362_050 |
0.1365237191 |
transcription factor |
TF Family: NOZZLE |
PREDICTED: uncharacterized protein LOC105635737 [Jatropha curcas] |
| 11 |
Hb_023956_010 |
0.1398140154 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 12 |
Hb_130967_010 |
0.1409317357 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_006846_030 |
0.1424059938 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 14 |
Hb_000483_130 |
0.1435303269 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002042_160 |
0.145315669 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 16 |
Hb_007632_130 |
0.1453872694 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000847_050 |
0.1479594385 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 18 |
Hb_002236_010 |
0.1484184478 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 19 |
Hb_000020_030 |
0.1494005457 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_009615_100 |
0.1504344048 |
- |
- |
hypothetical protein JCGZ_24459 [Jatropha curcas] |