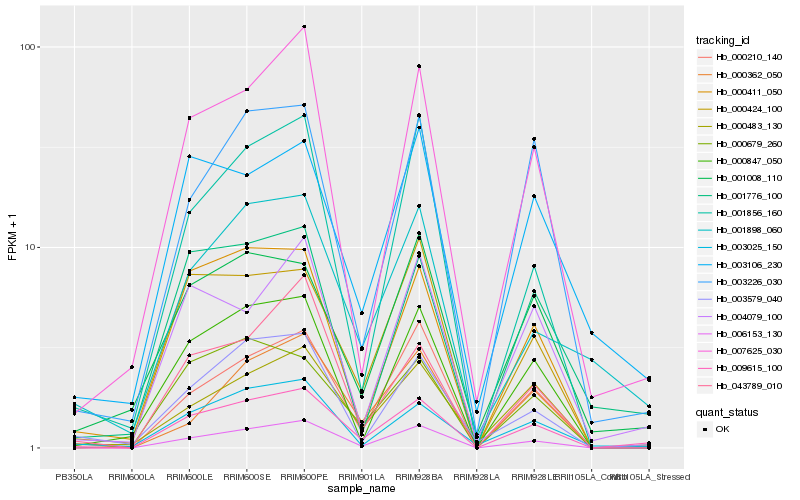
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009615_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_24459 [Jatropha curcas] |
| 2 |
Hb_006153_130 |
0.1249799923 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_000411_050 |
0.1399790157 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 4 |
Hb_007625_030 |
0.1455088445 |
- |
- |
unknown [Medicago truncatula] |
| 5 |
Hb_000210_140 |
0.1504344048 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 6 |
Hb_003025_150 |
0.1508234296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000847_050 |
0.1509920771 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 8 |
Hb_003579_040 |
0.1514700376 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 9 |
Hb_000424_100 |
0.1559941299 |
desease resistance |
Gene Name: G-alpha |
GTP-binding protein (p) alpha subunit, gpa1, putative [Ricinus communis] |
| 10 |
Hb_003226_030 |
0.1568620086 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 11 |
Hb_001776_100 |
0.1570385681 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000679_260 |
0.1585789232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004079_100 |
0.1588029928 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 14 |
Hb_000483_130 |
0.1602893578 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001008_110 |
0.161315158 |
- |
- |
- |
| 16 |
Hb_001898_060 |
0.1617185473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000362_050 |
0.1641157725 |
transcription factor |
TF Family: NOZZLE |
PREDICTED: uncharacterized protein LOC105635737 [Jatropha curcas] |
| 18 |
Hb_043789_010 |
0.1643427881 |
transcription factor |
TF Family: AP2 |
hypothetical protein VITISV_034131 [Vitis vinifera] |
| 19 |
Hb_001856_160 |
0.1655560581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003106_230 |
0.1659637404 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |