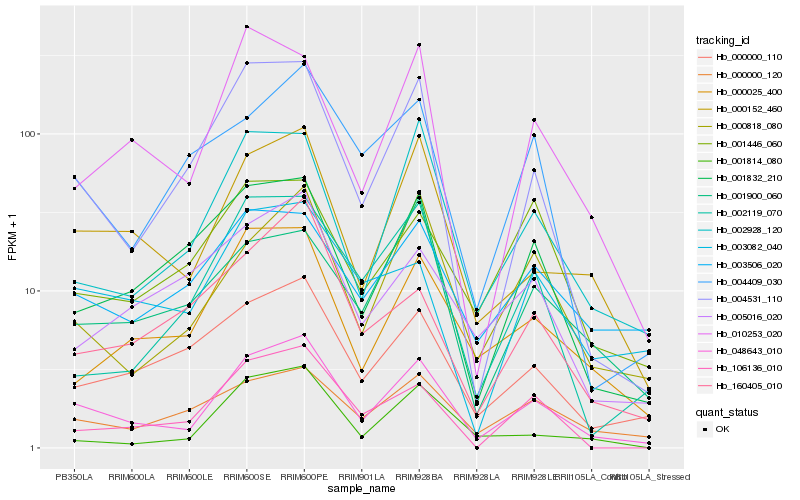
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048643_010 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_000152_460 |
0.1296738381 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 3 |
Hb_004531_110 |
0.1452697048 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 4 |
Hb_000818_080 |
0.1569512081 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 5 |
Hb_000000_110 |
0.1574585567 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 6 |
Hb_002928_120 |
0.1584040838 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 7 |
Hb_000025_400 |
0.1614308619 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 8 |
Hb_001832_210 |
0.1660503061 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 9 |
Hb_000000_120 |
0.1683820009 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 10 |
Hb_010253_020 |
0.1744447318 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_003082_040 |
0.1773530028 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Phoenix dactylifera] |
| 12 |
Hb_106136_010 |
0.1775580447 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 13 |
Hb_003506_020 |
0.1799225953 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 14 |
Hb_004409_030 |
0.1808047404 |
- |
- |
Glutamine synthetase 1,4 [Theobroma cacao] |
| 15 |
Hb_002119_070 |
0.186341066 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_001814_080 |
0.1868068806 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 17 |
Hb_005016_020 |
0.1869534171 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_160405_010 |
0.1875961806 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 19 |
Hb_001900_060 |
0.1899681733 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 20 |
Hb_001446_060 |
0.191769038 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |