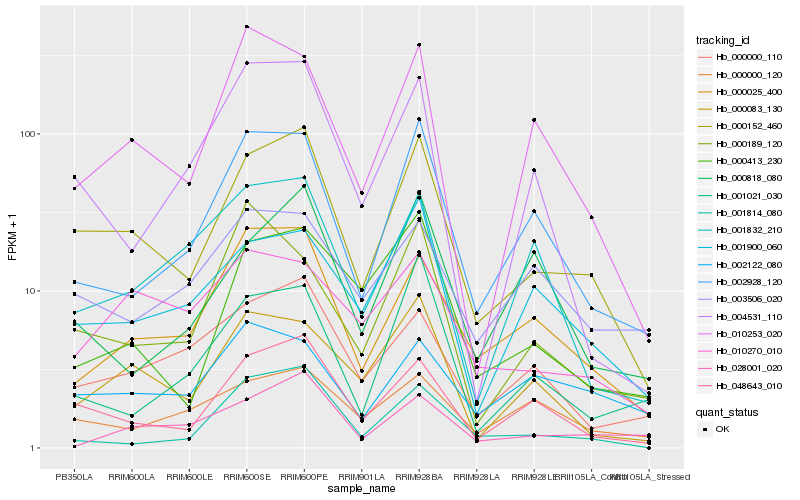
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_460 |
0.0 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 2 |
Hb_048643_010 |
0.1296738381 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_000025_400 |
0.1694411119 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 4 |
Hb_002928_120 |
0.1702920327 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 5 |
Hb_001900_060 |
0.1738523524 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 6 |
Hb_000083_130 |
0.178071903 |
- |
- |
hypothetical protein RCOM_0884570 [Ricinus communis] |
| 7 |
Hb_000000_110 |
0.1811013505 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 8 |
Hb_010253_020 |
0.1843321497 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_000413_230 |
0.1888952818 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 10 |
Hb_001814_080 |
0.1921376848 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 11 |
Hb_004531_110 |
0.1964328219 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 12 |
Hb_010270_010 |
0.200386348 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000818_080 |
0.2020414678 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 14 |
Hb_000000_120 |
0.2021484436 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 15 |
Hb_001832_210 |
0.2048299067 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 16 |
Hb_001021_030 |
0.2055572548 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_003506_020 |
0.208037291 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 18 |
Hb_002122_080 |
0.209324379 |
- |
- |
- |
| 19 |
Hb_028001_020 |
0.2098303329 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 20 |
Hb_000189_120 |
0.2134912684 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |