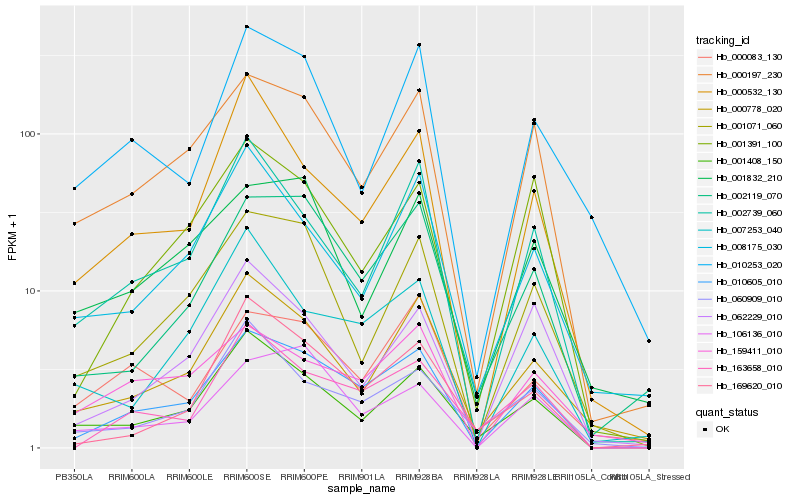
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010605_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 2 |
Hb_000197_230 |
0.1118505378 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 3 |
Hb_002119_070 |
0.134036991 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_163658_010 |
0.1458897037 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 5 |
Hb_001408_150 |
0.1480169479 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001071_060 |
0.1507407604 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 7 |
Hb_001391_100 |
0.1530470525 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 8 |
Hb_000778_020 |
0.154102488 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 9 |
Hb_159411_010 |
0.1586749915 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 10 |
Hb_002739_060 |
0.1590248015 |
- |
- |
- |
| 11 |
Hb_106136_010 |
0.1638740995 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 12 |
Hb_169620_010 |
0.165603361 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 13 |
Hb_062229_010 |
0.1684145264 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 14 |
Hb_000532_130 |
0.170045137 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 15 |
Hb_007253_040 |
0.1719106042 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000083_130 |
0.1730356446 |
- |
- |
hypothetical protein RCOM_0884570 [Ricinus communis] |
| 17 |
Hb_060909_010 |
0.1738757411 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 18 |
Hb_001832_210 |
0.174944495 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 19 |
Hb_008175_030 |
0.1755373387 |
transcription factor |
TF Family: TAZ |
PREDICTED: BTB/POZ and TAZ domain-containing protein 3 [Jatropha curcas] |
| 20 |
Hb_010253_020 |
0.179179425 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |