| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
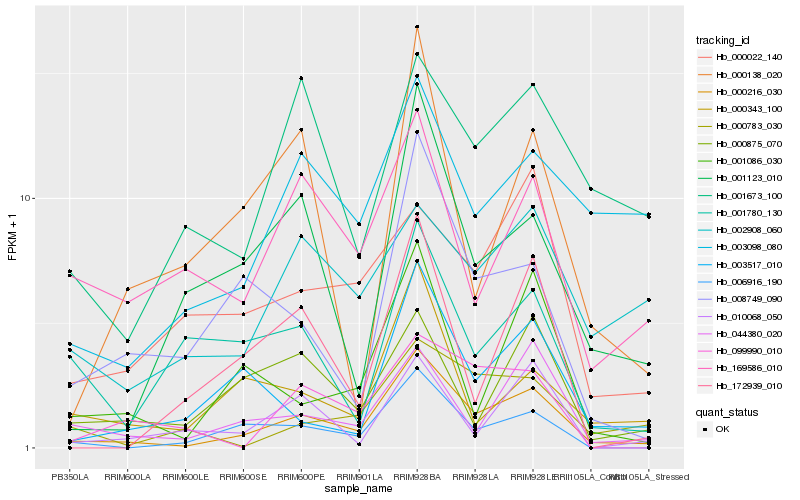
Hb_000216_030 |
0.0 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 2 |
Hb_006916_190 |
0.2022396234 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_044380_020 |
0.2175334774 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 4 |
Hb_001086_030 |
0.220475271 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 5 |
Hb_003517_010 |
0.2231272583 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_008749_090 |
0.2268452951 |
- |
- |
hypothetical protein CICLE_v10017161mg [Citrus clementina] |
| 7 |
Hb_001123_010 |
0.2354527574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000343_100 |
0.2379151942 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 9 |
Hb_000875_070 |
0.2419344678 |
- |
- |
- |
| 10 |
Hb_000783_030 |
0.247271057 |
- |
- |
tfiif-alpha, putative [Ricinus communis] |
| 11 |
Hb_003098_080 |
0.2476050248 |
- |
- |
- |
| 12 |
Hb_172939_010 |
0.250145398 |
- |
- |
unknown [Picea sitchensis] |
| 13 |
Hb_169586_010 |
0.2510920659 |
- |
- |
Acid phosphatase/vanadium-dependent haloperoxidase-related protein [Theobroma cacao] |
| 14 |
Hb_000022_140 |
0.2525659505 |
- |
- |
hypothetical protein MIMGU_mgv1a009920mg [Erythranthe guttata] |
| 15 |
Hb_010068_050 |
0.2539017582 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 16 |
Hb_099990_010 |
0.2590348273 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 11 [Vitis vinifera] |
| 17 |
Hb_001780_130 |
0.259919684 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 18 |
Hb_001673_100 |
0.2602751685 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 19 |
Hb_000138_020 |
0.2604296473 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 20 |
Hb_002908_060 |
0.2611318753 |
- |
- |
unnamed protein product, partial [Coffea canephora] |