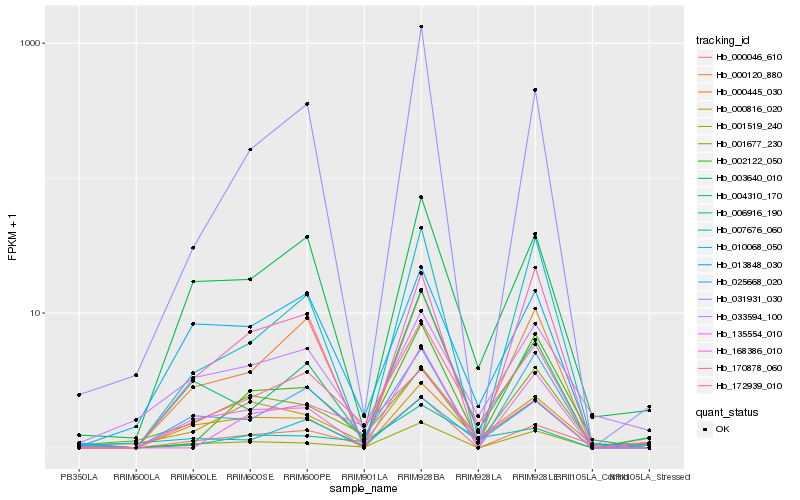
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172939_010 |
0.0 |
- |
- |
unknown [Picea sitchensis] |
| 2 |
Hb_000120_880 |
0.1509737872 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 3 |
Hb_004310_170 |
0.1734724809 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 4 |
Hb_025668_020 |
0.1740977584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003640_010 |
0.1748922591 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 6 |
Hb_002122_050 |
0.1796417529 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 7 |
Hb_168386_010 |
0.1806168691 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 8 |
Hb_010068_050 |
0.1811875479 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 9 |
Hb_000445_030 |
0.1817564564 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 10 |
Hb_001519_240 |
0.1825343231 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_000816_020 |
0.1888633362 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_007676_060 |
0.1939408775 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 13 |
Hb_001677_230 |
0.1943554068 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000046_610 |
0.1977192343 |
- |
- |
PREDICTED: putative cullin-like protein 2 isoform X3 [Populus euphratica] |
| 15 |
Hb_135554_010 |
0.1983668512 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 16 |
Hb_013848_030 |
0.199878822 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 17 |
Hb_006916_190 |
0.2005024367 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_170878_060 |
0.2068703734 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 19 |
Hb_033594_100 |
0.2075375478 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_031931_030 |
0.2090286035 |
- |
- |
lipid binding protein, putative [Ricinus communis] |