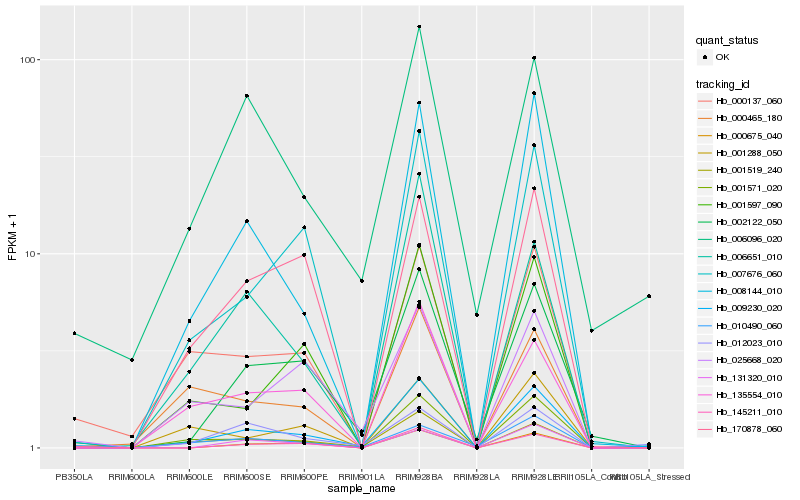
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009230_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008144_010 |
0.1396735012 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 3 |
Hb_001519_240 |
0.1436748904 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 4 |
Hb_001571_020 |
0.1473559162 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850-like [Citrus sinensis] |
| 5 |
Hb_002122_050 |
0.1476215009 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 6 |
Hb_007676_060 |
0.1561926843 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 7 |
Hb_000137_060 |
0.1628097909 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 8 |
Hb_000675_040 |
0.1710575613 |
- |
- |
hypothetical protein OsI_02937 [Oryza sativa Indica Group] |
| 9 |
Hb_145211_010 |
0.1716567427 |
- |
- |
hypothetical protein M569_05648, partial [Genlisea aurea] |
| 10 |
Hb_001597_090 |
0.1734774609 |
- |
- |
- |
| 11 |
Hb_012023_010 |
0.1751243665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006651_010 |
0.1770973804 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4-like [Citrus sinensis] |
| 13 |
Hb_135554_010 |
0.1776091814 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 14 |
Hb_025668_020 |
0.1810543796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006096_020 |
0.1811687034 |
- |
- |
Os06g0343500 [Oryza sativa Japonica Group] |
| 16 |
Hb_010490_060 |
0.1848645656 |
- |
- |
PREDICTED: uncharacterized protein LOC105767021 [Gossypium raimondii] |
| 17 |
Hb_131320_010 |
0.1901319386 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000465_180 |
0.1917768733 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 19 |
Hb_170878_060 |
0.1921486317 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 20 |
Hb_001288_050 |
0.1955025026 |
- |
- |
- |