| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
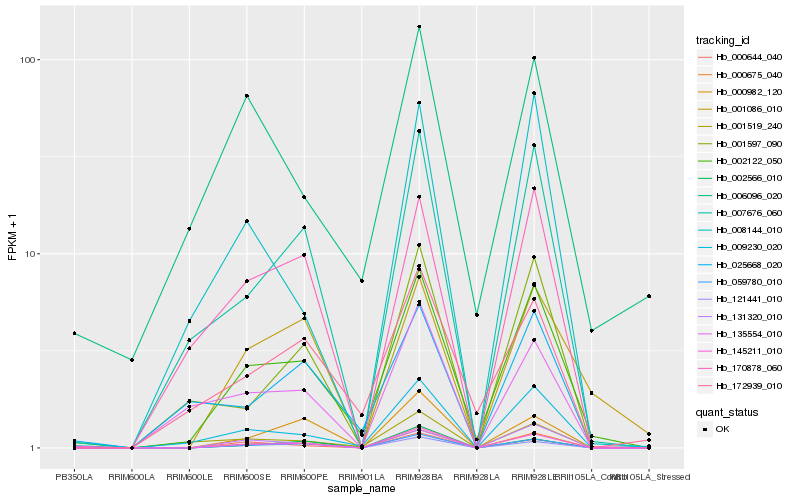
Hb_002122_050 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 2 |
Hb_000675_040 |
0.129759595 |
- |
- |
hypothetical protein OsI_02937 [Oryza sativa Indica Group] |
| 3 |
Hb_145211_010 |
0.1385854649 |
- |
- |
hypothetical protein M569_05648, partial [Genlisea aurea] |
| 4 |
Hb_059780_010 |
0.1456813702 |
- |
- |
PREDICTED: uncharacterized protein LOC104885482, partial [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_009230_020 |
0.1476215009 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007676_060 |
0.1541085336 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 7 |
Hb_131320_010 |
0.1731244832 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 8 |
Hb_121441_010 |
0.1743649873 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_172939_010 |
0.1796417529 |
- |
- |
unknown [Picea sitchensis] |
| 10 |
Hb_170878_060 |
0.1814348209 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 11 |
Hb_000644_040 |
0.1861990559 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_009157 [Vitis vinifera] |
| 12 |
Hb_001519_240 |
0.1869279738 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_025668_020 |
0.1966757879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006096_020 |
0.1970972763 |
- |
- |
Os06g0343500 [Oryza sativa Japonica Group] |
| 15 |
Hb_001597_090 |
0.1976148604 |
- |
- |
- |
| 16 |
Hb_001086_010 |
0.2022186507 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 17 |
Hb_008144_010 |
0.2038255617 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 18 |
Hb_135554_010 |
0.2041329926 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 19 |
Hb_002566_010 |
0.2049769159 |
- |
- |
PREDICTED: uncharacterized protein LOC105643984 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000982_120 |
0.2052967207 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |