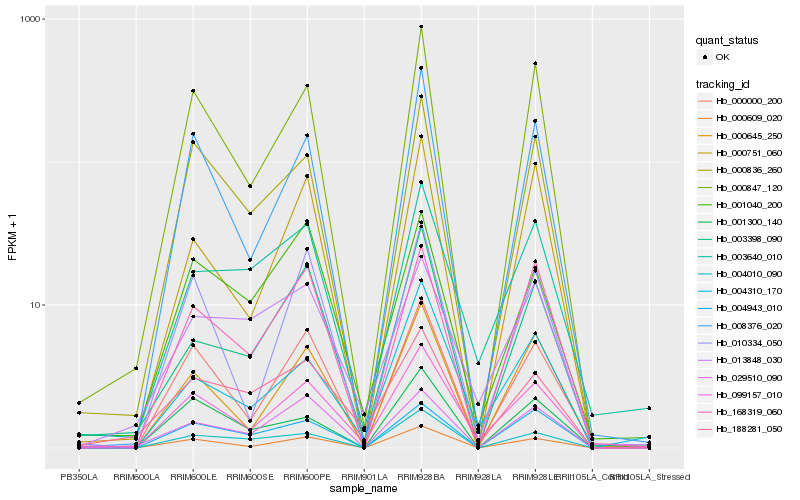
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_099157_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 2 |
Hb_000000_200 |
0.1100826913 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 [Jatropha curcas] |
| 3 |
Hb_000609_020 |
0.1109807622 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 4 |
Hb_008376_020 |
0.1182824752 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 5 |
Hb_000847_120 |
0.11921323 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 6 |
Hb_000645_250 |
0.1316729791 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 7 |
Hb_000836_260 |
0.1334352716 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_004310_170 |
0.1418939916 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 9 |
Hb_010334_050 |
0.1434145322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003640_010 |
0.1497615001 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 11 |
Hb_168319_060 |
0.1513032531 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 12 |
Hb_004010_090 |
0.1545761884 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_188281_050 |
0.1571999284 |
- |
- |
PREDICTED: solute carrier family 35 member F1-like isoform X1 [Vitis vinifera] |
| 14 |
Hb_001040_200 |
0.1573482928 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 15 |
Hb_000751_060 |
0.1586266564 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 16 |
Hb_013848_030 |
0.1618809329 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 17 |
Hb_001300_140 |
0.1625934063 |
- |
- |
- |
| 18 |
Hb_004943_010 |
0.163414227 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_003398_090 |
0.1640029013 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 20 |
Hb_029510_090 |
0.1661511214 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |