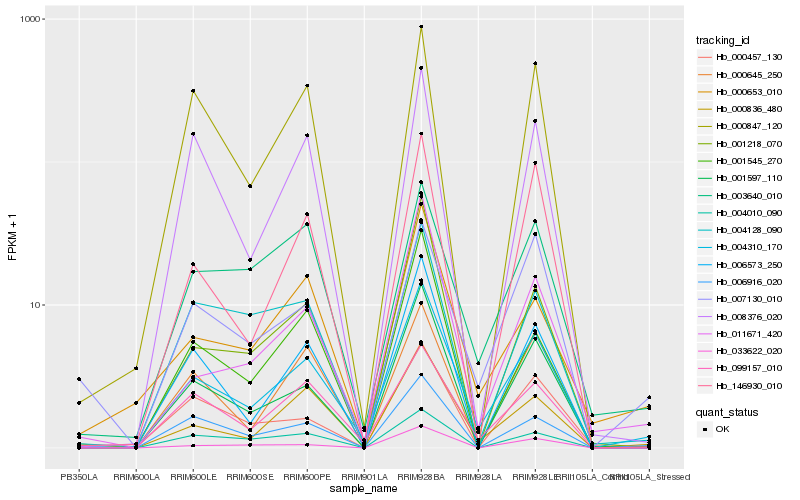
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004310_170 |
0.0 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 2 |
Hb_000836_480 |
0.1320767759 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_001597_110 |
0.1362477952 |
- |
- |
- |
| 4 |
Hb_006573_250 |
0.1417364201 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 5 |
Hb_099157_010 |
0.1418939916 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 6 |
Hb_006916_020 |
0.1493907802 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 7 |
Hb_000653_010 |
0.1565979113 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 8 |
Hb_146930_010 |
0.1582716689 |
- |
- |
- |
| 9 |
Hb_011671_420 |
0.1584576677 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 10 |
Hb_000645_250 |
0.1589010307 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 11 |
Hb_033622_020 |
0.1599748875 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 12 |
Hb_008376_020 |
0.1609571588 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 13 |
Hb_001218_070 |
0.1626295059 |
- |
- |
PREDICTED: sugar transport protein 5-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003640_010 |
0.1628396791 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 15 |
Hb_004128_090 |
0.163564798 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 16 |
Hb_000847_120 |
0.16646954 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 17 |
Hb_001545_270 |
0.1673158027 |
- |
- |
laccase, putative [Ricinus communis] |
| 18 |
Hb_000457_130 |
0.1685837177 |
- |
- |
hypothetical protein AALP_AA3G009100 [Arabis alpina] |
| 19 |
Hb_004010_090 |
0.1690882549 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_007130_010 |
0.1714028149 |
- |
- |
- |