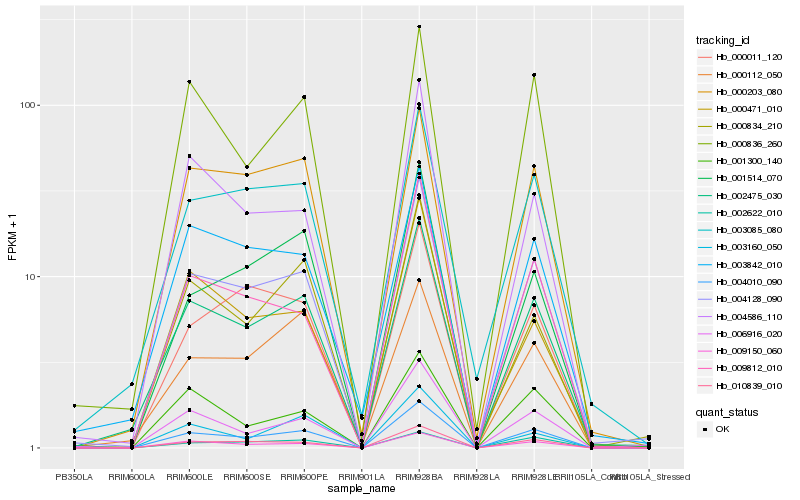
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004010_090 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_004128_090 |
0.0589529737 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 3 |
Hb_002475_030 |
0.0711597147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010839_010 |
0.0733901085 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 5 |
Hb_009150_060 |
0.0783787025 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 6 |
Hb_009812_010 |
0.1083149978 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_001514_070 |
0.1131185605 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000836_260 |
0.1136655879 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 9 |
Hb_000834_210 |
0.1144168105 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_006916_020 |
0.1161834208 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 11 |
Hb_001300_140 |
0.1172225382 |
- |
- |
- |
| 12 |
Hb_004586_110 |
0.1173177106 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 13 |
Hb_003085_080 |
0.1232217596 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_002622_010 |
0.124503689 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 15 |
Hb_000011_120 |
0.1261352996 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 16 |
Hb_000203_080 |
0.1282497547 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 17 |
Hb_003160_050 |
0.1283796999 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 18 |
Hb_000471_010 |
0.1316223645 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_003842_010 |
0.13254511 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000112_050 |
0.1325690444 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |