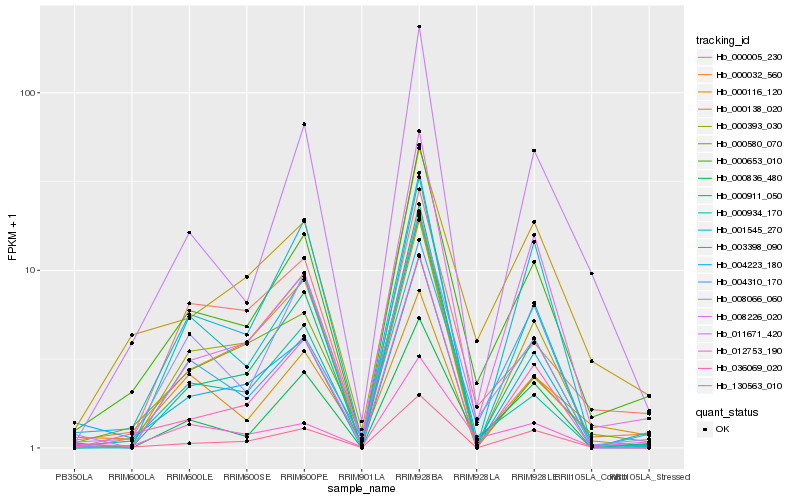
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000653_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 2 |
Hb_008066_060 |
0.1220598098 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 3 |
Hb_130563_010 |
0.1315554239 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_012753_190 |
0.1347390358 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 5 |
Hb_000836_480 |
0.1352859985 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_008226_020 |
0.1396199232 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 7 |
Hb_000138_020 |
0.1436857499 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 8 |
Hb_000911_050 |
0.1438242472 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 21 [Jatropha curcas] |
| 9 |
Hb_036069_020 |
0.151359191 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 10 |
Hb_004223_180 |
0.1556916255 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 11 |
Hb_004310_170 |
0.1565979113 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 12 |
Hb_000032_560 |
0.1568983565 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 13 |
Hb_001545_270 |
0.1591878871 |
- |
- |
laccase, putative [Ricinus communis] |
| 14 |
Hb_000005_230 |
0.1621850368 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 15 |
Hb_000580_070 |
0.1634577662 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 16 |
Hb_000116_120 |
0.1636902309 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 17 |
Hb_000393_030 |
0.1645512482 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_011671_420 |
0.1677794626 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 19 |
Hb_003398_090 |
0.1715488072 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 20 |
Hb_000934_170 |
0.1722169215 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |