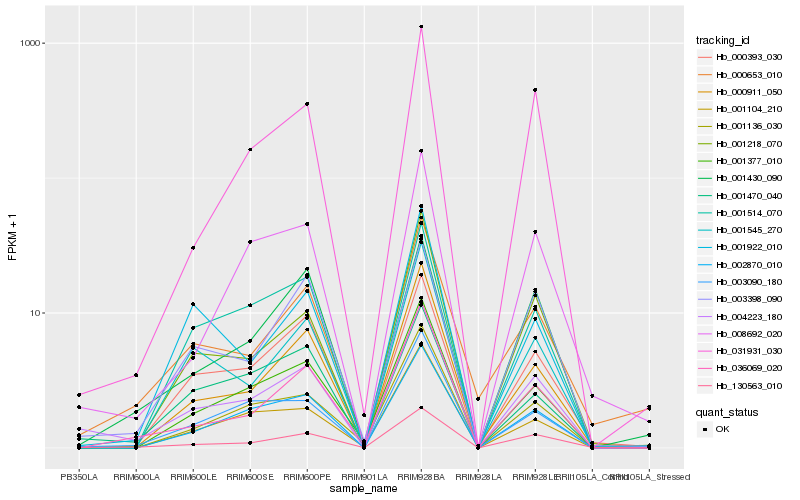
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_180 |
0.0 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 2 |
Hb_130563_010 |
0.099053599 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_036069_020 |
0.1350156515 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 4 |
Hb_003090_180 |
0.1370677283 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 5 |
Hb_002870_010 |
0.1379028225 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 6 |
Hb_001377_010 |
0.141371936 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 7 |
Hb_000911_050 |
0.1461438228 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 21 [Jatropha curcas] |
| 8 |
Hb_001104_210 |
0.146231047 |
- |
- |
hypothetical protein JCGZ_23620 [Jatropha curcas] |
| 9 |
Hb_001218_070 |
0.1471137639 |
- |
- |
PREDICTED: sugar transport protein 5-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_008692_020 |
0.1485723562 |
- |
- |
- |
| 11 |
Hb_001470_040 |
0.1491161116 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 12 |
Hb_000393_030 |
0.1494192177 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001514_070 |
0.1499781744 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_001545_270 |
0.1501789414 |
- |
- |
laccase, putative [Ricinus communis] |
| 15 |
Hb_003398_090 |
0.1513409064 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 16 |
Hb_001136_030 |
0.1517983167 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25-like [Jatropha curcas] |
| 17 |
Hb_031931_030 |
0.1543886851 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000653_010 |
0.1556916255 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 19 |
Hb_001430_090 |
0.1648135035 |
- |
- |
hypothetical protein JCGZ_05131 [Jatropha curcas] |
| 20 |
Hb_001922_010 |
0.1669368647 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |