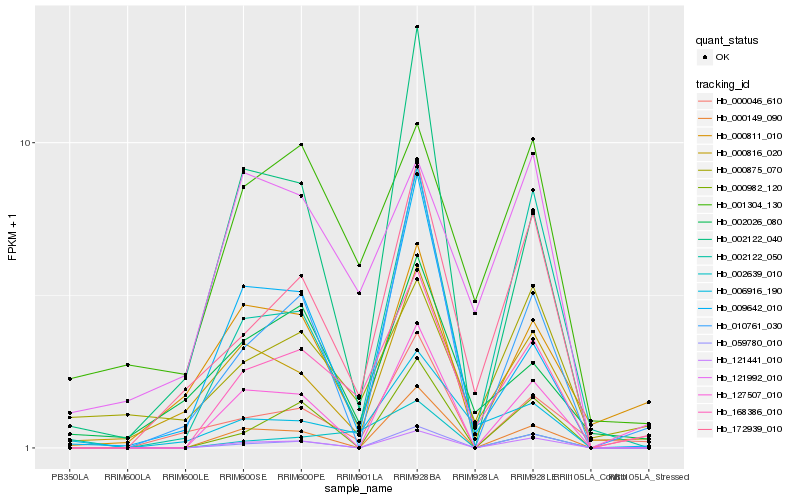
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168386_010 |
0.0 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 2 |
Hb_172939_010 |
0.1806168691 |
- |
- |
unknown [Picea sitchensis] |
| 3 |
Hb_006916_190 |
0.221013933 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_002639_010 |
0.2307751434 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 5 |
Hb_001304_130 |
0.2325587168 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like transcription factor PAT1 [Jatropha curcas] |
| 6 |
Hb_009642_010 |
0.2369598553 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 7 |
Hb_121441_010 |
0.2395488141 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 8 |
Hb_000816_020 |
0.2406048384 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_127507_010 |
0.243408362 |
- |
- |
PREDICTED: uncharacterized protein LOC104814513 [Tarenaya hassleriana] |
| 10 |
Hb_002122_050 |
0.2450547189 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_000046_610 |
0.2492785608 |
- |
- |
PREDICTED: putative cullin-like protein 2 isoform X3 [Populus euphratica] |
| 12 |
Hb_010761_030 |
0.2517815938 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_121992_010 |
0.2519199941 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 14 |
Hb_000149_090 |
0.2519730022 |
- |
- |
- |
| 15 |
Hb_000875_070 |
0.2535570266 |
- |
- |
- |
| 16 |
Hb_002122_040 |
0.2554628838 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_000982_120 |
0.2573887817 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |
| 18 |
Hb_000811_010 |
0.257544279 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 19 |
Hb_002026_080 |
0.2577679884 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 20 |
Hb_059780_010 |
0.2578586853 |
- |
- |
PREDICTED: uncharacterized protein LOC104885482, partial [Beta vulgaris subsp. vulgaris] |