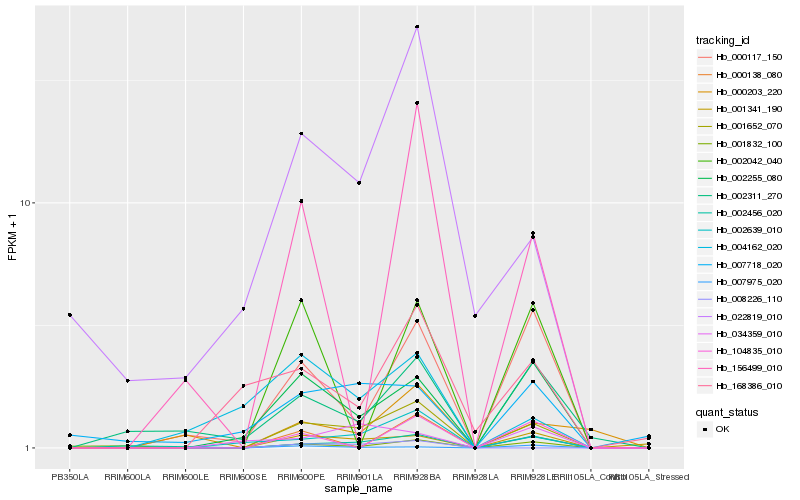
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001652_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103439285 [Malus domestica] |
| 2 |
Hb_002639_010 |
0.2290247914 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 3 |
Hb_002255_080 |
0.2303589715 |
- |
- |
- |
| 4 |
Hb_001341_190 |
0.2558106242 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002456_020 |
0.2574659739 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 6 |
Hb_000117_150 |
0.2684580197 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 7 |
Hb_022819_010 |
0.2869588804 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 8 |
Hb_000138_080 |
0.2992826184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007975_020 |
0.3015103968 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_002311_270 |
0.3056029432 |
- |
- |
PREDICTED: WD repeat-containing protein 43-like [Malus domestica] |
| 11 |
Hb_168386_010 |
0.3065644927 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 12 |
Hb_104835_010 |
0.3129212082 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 13 |
Hb_156499_010 |
0.320404753 |
- |
- |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 14 |
Hb_001832_100 |
0.3204573462 |
- |
- |
- |
| 15 |
Hb_000203_220 |
0.3220131026 |
- |
- |
PREDICTED: uncharacterized protein LOC105168575 [Sesamum indicum] |
| 16 |
Hb_007718_020 |
0.3227587555 |
- |
- |
PREDICTED: receptor-like protein 12 [Camelina sativa] |
| 17 |
Hb_002042_040 |
0.3262437507 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 18 |
Hb_004162_020 |
0.3306846753 |
- |
- |
- |
| 19 |
Hb_008226_110 |
0.3316593541 |
- |
- |
uncharacterized protein LOC100501343 [Zea mays] |
| 20 |
Hb_034359_010 |
0.333339257 |
- |
- |
- |