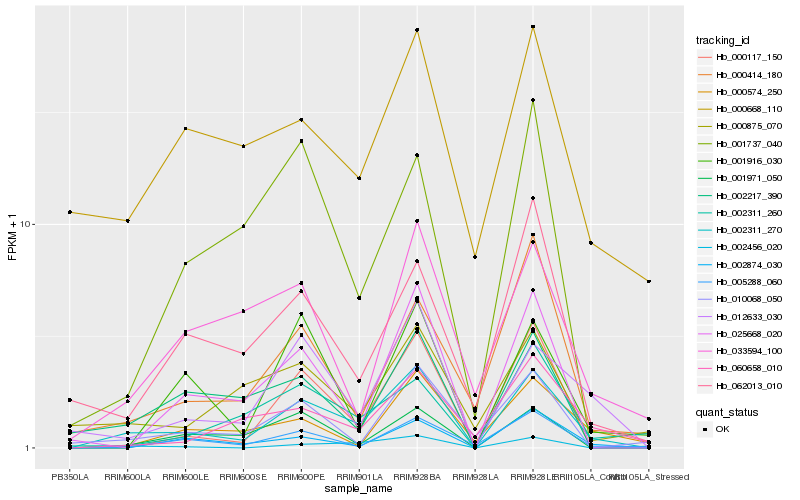
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_270 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein 43-like [Malus domestica] |
| 2 |
Hb_002456_020 |
0.1984244619 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 3 |
Hb_012633_030 |
0.2078405543 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 4 |
Hb_000117_150 |
0.216915488 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 5 |
Hb_005288_060 |
0.2252071687 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 6 |
Hb_001971_050 |
0.2313214339 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_002874_030 |
0.2350106129 |
- |
- |
hypothetical protein JCGZ_07001 [Jatropha curcas] |
| 8 |
Hb_025668_020 |
0.2364688973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002217_390 |
0.2390720912 |
- |
- |
PREDICTED: galactokinase [Jatropha curcas] |
| 10 |
Hb_002311_260 |
0.2464248037 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 11 |
Hb_060658_010 |
0.248656082 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 12 |
Hb_062013_010 |
0.2490648862 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 13 |
Hb_001916_030 |
0.2512985628 |
- |
- |
PREDICTED: uncharacterized protein LOC105802975 [Gossypium raimondii] |
| 14 |
Hb_000414_180 |
0.2519721938 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 15 |
Hb_010068_050 |
0.2555118523 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 16 |
Hb_001737_040 |
0.2568751334 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 17 |
Hb_000668_110 |
0.2584356218 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 18 |
Hb_033594_100 |
0.2589364591 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000875_070 |
0.2590216616 |
- |
- |
- |
| 20 |
Hb_000574_250 |
0.2613795952 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |