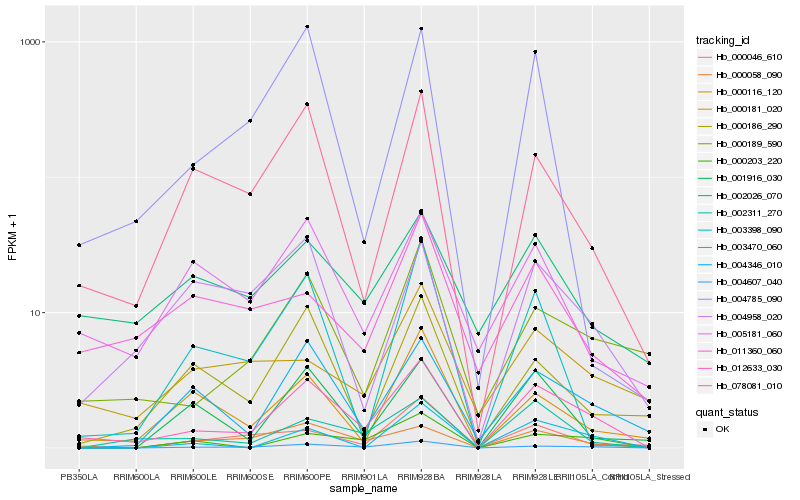
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012633_030 |
0.0 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 2 |
Hb_004607_040 |
0.1708149536 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 3 |
Hb_000058_090 |
0.2021506338 |
- |
- |
- |
| 4 |
Hb_002311_270 |
0.2078405543 |
- |
- |
PREDICTED: WD repeat-containing protein 43-like [Malus domestica] |
| 5 |
Hb_078081_010 |
0.2093458468 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 6 |
Hb_000189_590 |
0.2126461242 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 7 |
Hb_004346_010 |
0.2222453843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000203_220 |
0.2289937539 |
- |
- |
PREDICTED: uncharacterized protein LOC105168575 [Sesamum indicum] |
| 9 |
Hb_005181_060 |
0.2390001709 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 10 |
Hb_004785_090 |
0.2426550326 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 11 |
Hb_002026_070 |
0.2439308532 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 12 |
Hb_003470_060 |
0.2450172348 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 45 [Jatropha curcas] |
| 13 |
Hb_000116_120 |
0.24517619 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 14 |
Hb_001916_030 |
0.2456687763 |
- |
- |
PREDICTED: uncharacterized protein LOC105802975 [Gossypium raimondii] |
| 15 |
Hb_004958_020 |
0.2491951834 |
- |
- |
PREDICTED: uncharacterized protein LOC104609039 [Nelumbo nucifera] |
| 16 |
Hb_000186_290 |
0.251478678 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003398_090 |
0.2553495908 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 18 |
Hb_000181_020 |
0.2560610013 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_011360_060 |
0.2579341099 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 20 |
Hb_000046_610 |
0.2587279178 |
- |
- |
PREDICTED: putative cullin-like protein 2 isoform X3 [Populus euphratica] |