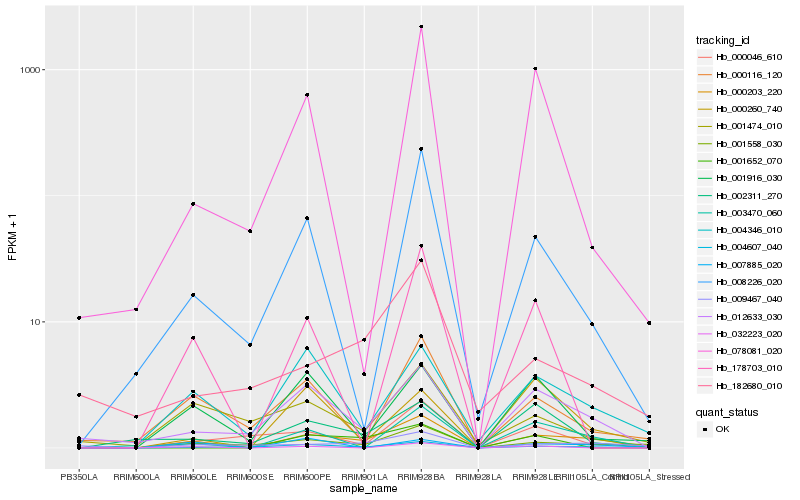
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000203_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105168575 [Sesamum indicum] |
| 2 |
Hb_004607_040 |
0.1984002476 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 3 |
Hb_012633_030 |
0.2289937539 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 4 |
Hb_003470_060 |
0.2347271682 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 45 [Jatropha curcas] |
| 5 |
Hb_001558_030 |
0.253466602 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 6 |
Hb_004346_010 |
0.2578200149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002311_270 |
0.2830592378 |
- |
- |
PREDICTED: WD repeat-containing protein 43-like [Malus domestica] |
| 8 |
Hb_008226_020 |
0.2878296478 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 9 |
Hb_000116_120 |
0.2903110301 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 10 |
Hb_001916_030 |
0.2930433952 |
- |
- |
PREDICTED: uncharacterized protein LOC105802975 [Gossypium raimondii] |
| 11 |
Hb_009467_040 |
0.3001536248 |
- |
- |
PREDICTED: putative B3 domain-containing protein At3g24850 [Jatropha curcas] |
| 12 |
Hb_001474_010 |
0.30316801 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000046_610 |
0.305619921 |
- |
- |
PREDICTED: putative cullin-like protein 2 isoform X3 [Populus euphratica] |
| 14 |
Hb_178703_010 |
0.3173103905 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 15 |
Hb_032223_020 |
0.3180711252 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_182680_010 |
0.3183610128 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 17 |
Hb_007885_020 |
0.3194900858 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 18 |
Hb_001652_070 |
0.3220131026 |
- |
- |
PREDICTED: uncharacterized protein LOC103439285 [Malus domestica] |
| 19 |
Hb_000260_740 |
0.3221617156 |
- |
- |
- |
| 20 |
Hb_078081_020 |
0.3229447925 |
- |
- |
- |