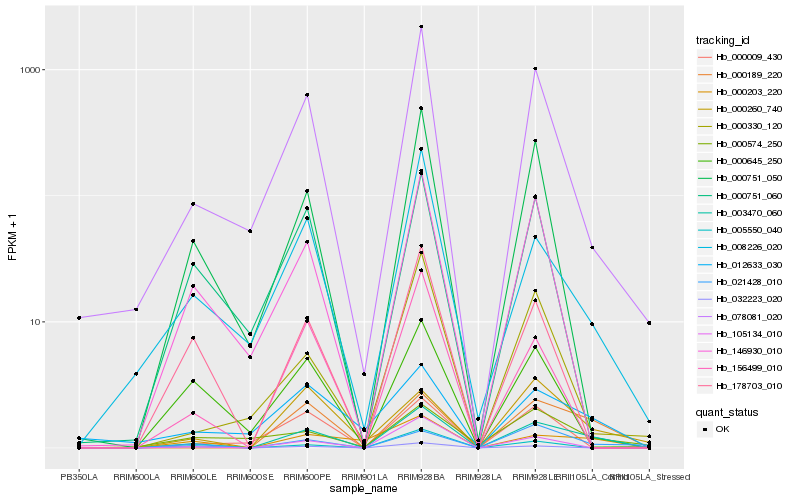
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003470_060 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 45 [Jatropha curcas] |
| 2 |
Hb_000189_220 |
0.1843818263 |
- |
- |
- |
| 3 |
Hb_078081_020 |
0.2030316882 |
- |
- |
- |
| 4 |
Hb_008226_020 |
0.2297866045 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 5 |
Hb_000751_050 |
0.2332472922 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 6 |
Hb_000203_220 |
0.2347271682 |
- |
- |
PREDICTED: uncharacterized protein LOC105168575 [Sesamum indicum] |
| 7 |
Hb_146930_010 |
0.2424050047 |
- |
- |
- |
| 8 |
Hb_000260_740 |
0.2426805329 |
- |
- |
- |
| 9 |
Hb_012633_030 |
0.2450172348 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 10 |
Hb_021428_010 |
0.2459769894 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 11 |
Hb_178703_010 |
0.2498221392 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 12 |
Hb_156499_010 |
0.2498601686 |
- |
- |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 13 |
Hb_105134_010 |
0.2525465389 |
- |
- |
hypothetical protein JCGZ_16347 [Jatropha curcas] |
| 14 |
Hb_000751_060 |
0.2582359007 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 15 |
Hb_005550_040 |
0.2593251412 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 16 |
Hb_032223_020 |
0.2606999181 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_000330_120 |
0.2615679542 |
- |
- |
PREDICTED: bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase-like [Nelumbo nucifera] |
| 18 |
Hb_000574_250 |
0.2622465999 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 19 |
Hb_000645_250 |
0.2626113158 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 20 |
Hb_000009_430 |
0.26528299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |