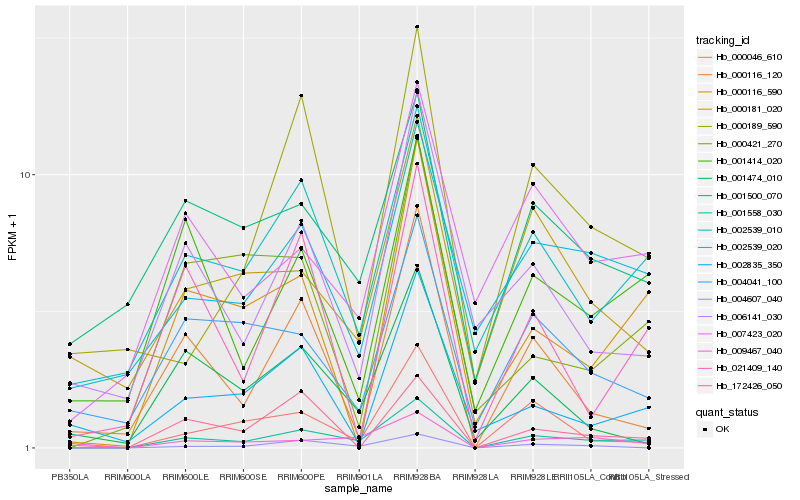
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001558_030 |
0.0 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 2 |
Hb_000046_610 |
0.1969874929 |
- |
- |
PREDICTED: putative cullin-like protein 2 isoform X3 [Populus euphratica] |
| 3 |
Hb_004607_040 |
0.2056788571 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 4 |
Hb_002539_010 |
0.2057934595 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 5 |
Hb_009467_040 |
0.2139465972 |
- |
- |
PREDICTED: putative B3 domain-containing protein At3g24850 [Jatropha curcas] |
| 6 |
Hb_000116_590 |
0.2153300345 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 7 |
Hb_000116_120 |
0.2210469742 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 8 |
Hb_001414_020 |
0.2220720851 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_007423_020 |
0.223958101 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 10 |
Hb_001474_010 |
0.2294120654 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002539_020 |
0.2295539476 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 12 |
Hb_006141_030 |
0.2305193228 |
- |
- |
- |
| 13 |
Hb_004041_100 |
0.2333950605 |
- |
- |
- |
| 14 |
Hb_000181_020 |
0.234829863 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000189_590 |
0.2352191681 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 16 |
Hb_021409_140 |
0.2361587397 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 17 |
Hb_172426_050 |
0.2385025937 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 18 |
Hb_001500_070 |
0.2393232297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000421_270 |
0.2404909961 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 20 |
Hb_002835_350 |
0.2417647335 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |