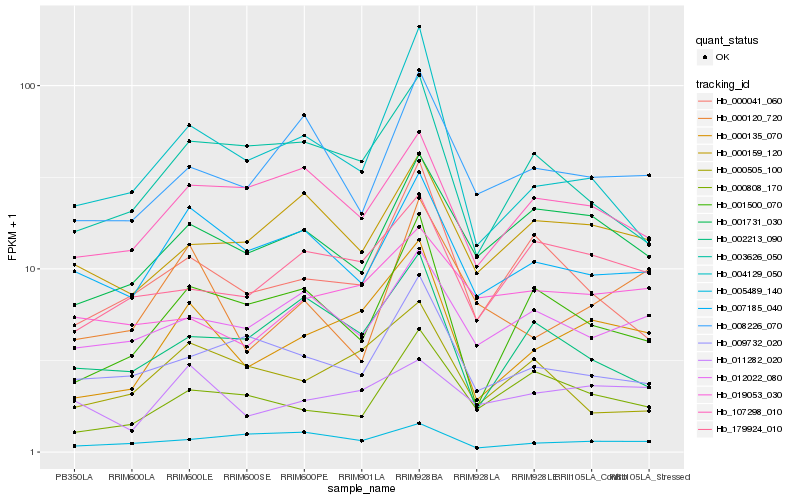
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000135_070 |
0.0 |
- |
- |
PREDICTED: inositol oxygenase 1-like [Jatropha curcas] |
| 2 |
Hb_179924_010 |
0.1741145175 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_001500_070 |
0.1745711651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004129_050 |
0.1817879828 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |
| 5 |
Hb_001731_030 |
0.1829935753 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A-like [Gossypium raimondii] |
| 6 |
Hb_007185_040 |
0.1888066741 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 7 |
Hb_003626_050 |
0.1897180217 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 8 |
Hb_107298_010 |
0.190365069 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 9 |
Hb_011282_020 |
0.1913049527 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000159_120 |
0.1927112187 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 11 |
Hb_000041_060 |
0.1931980698 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 12 |
Hb_000808_170 |
0.197026057 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 13 |
Hb_008226_070 |
0.1980125784 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 14 |
Hb_000505_100 |
0.1999711969 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC100250663 [Vitis vinifera] |
| 15 |
Hb_012022_080 |
0.2000663222 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_002213_090 |
0.2004004384 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 17 |
Hb_009732_020 |
0.2042865974 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 18 |
Hb_005489_140 |
0.205051618 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 19 |
Hb_000120_720 |
0.2060634121 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 20 |
Hb_019053_030 |
0.2061339981 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |