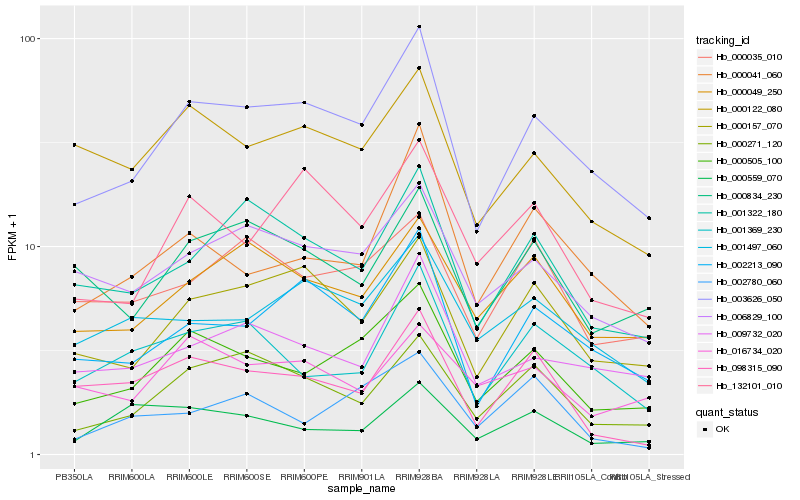
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000505_100 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC100250663 [Vitis vinifera] |
| 2 |
Hb_003626_050 |
0.1047590698 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 3 |
Hb_006829_100 |
0.1379206607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000122_080 |
0.1417325503 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 5 |
Hb_000041_060 |
0.1442705654 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 6 |
Hb_000035_010 |
0.1478433278 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000049_250 |
0.1483401337 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001369_230 |
0.1505022789 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 9 |
Hb_002780_060 |
0.1540253116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000271_120 |
0.1555309446 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 11 |
Hb_132101_010 |
0.1569014435 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 12 |
Hb_001497_060 |
0.1574744455 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 13 |
Hb_016734_020 |
0.1593092352 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_000834_230 |
0.1595784522 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 15 |
Hb_001322_180 |
0.1597125898 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 16 |
Hb_009732_020 |
0.159715299 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 17 |
Hb_000157_070 |
0.1602363128 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 18 |
Hb_002213_090 |
0.164452922 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 19 |
Hb_098315_090 |
0.1644731352 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 20 |
Hb_000559_070 |
0.1650507342 |
- |
- |
calcineurin B, putative [Ricinus communis] |