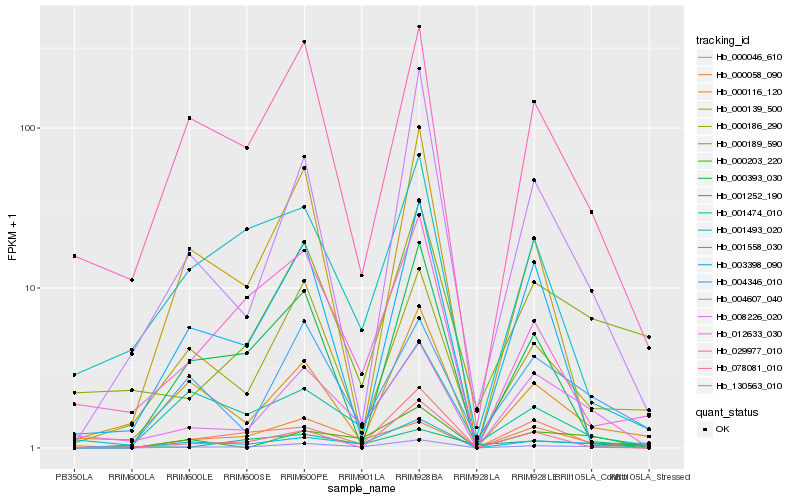
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004607_040 |
0.0 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 2 |
Hb_012633_030 |
0.1708149536 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 3 |
Hb_000203_220 |
0.1984002476 |
- |
- |
PREDICTED: uncharacterized protein LOC105168575 [Sesamum indicum] |
| 4 |
Hb_001558_030 |
0.2056788571 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 5 |
Hb_078081_010 |
0.2094894484 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 6 |
Hb_000046_610 |
0.2142198049 |
- |
- |
PREDICTED: putative cullin-like protein 2 isoform X3 [Populus euphratica] |
| 7 |
Hb_004346_010 |
0.2198088557 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000058_090 |
0.2208304508 |
- |
- |
- |
| 9 |
Hb_001474_010 |
0.2331139704 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000186_290 |
0.2332913112 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 11 |
Hb_029977_010 |
0.2333384609 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_000116_120 |
0.2346875555 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 13 |
Hb_000393_030 |
0.2405869445 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_008226_020 |
0.2410252198 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 15 |
Hb_000189_590 |
0.2412209855 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 16 |
Hb_001493_020 |
0.2492923033 |
- |
- |
leucine-rich repeat receptor-like protein kinase [Manihot esculenta] |
| 17 |
Hb_130563_010 |
0.2498759723 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001252_190 |
0.2514052619 |
desease resistance |
Gene Name: NB-ARC |
- |
| 19 |
Hb_003398_090 |
0.2519385857 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 20 |
Hb_000139_500 |
0.2536592803 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0286901 [Prunus mume] |