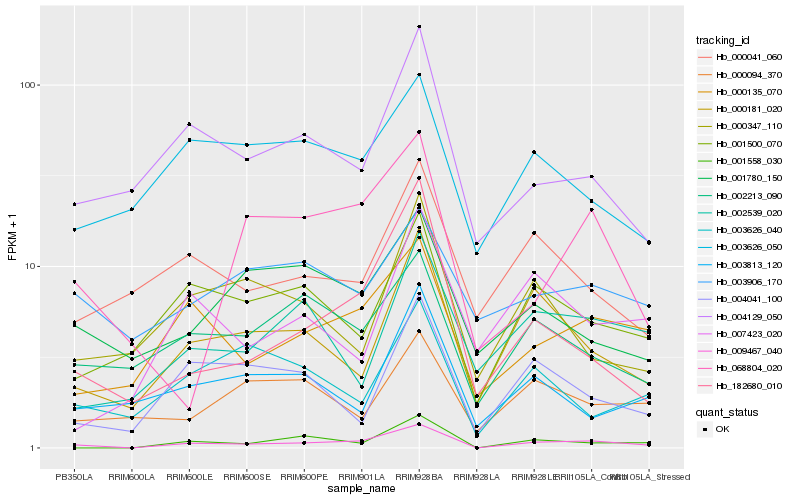
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009467_040 |
0.0 |
- |
- |
PREDICTED: putative B3 domain-containing protein At3g24850 [Jatropha curcas] |
| 2 |
Hb_000135_070 |
0.2125767175 |
- |
- |
PREDICTED: inositol oxygenase 1-like [Jatropha curcas] |
| 3 |
Hb_001558_030 |
0.2139465972 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 4 |
Hb_000181_020 |
0.2206959387 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_182680_010 |
0.2262954383 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 6 |
Hb_001500_070 |
0.2341144685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004129_050 |
0.2371524494 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |
| 8 |
Hb_002213_090 |
0.2406134211 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 9 |
Hb_004041_100 |
0.2464569574 |
- |
- |
- |
| 10 |
Hb_000041_060 |
0.2542627073 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_002539_020 |
0.2575537969 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 12 |
Hb_068804_020 |
0.258139845 |
- |
- |
hypothetical protein AMTR_s00046p00231970 [Amborella trichopoda] |
| 13 |
Hb_003626_050 |
0.2605915874 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 14 |
Hb_001780_150 |
0.2612685326 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 15 |
Hb_003813_120 |
0.2639338358 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_003626_040 |
0.2655798974 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000347_110 |
0.2663777606 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 18 |
Hb_000094_370 |
0.2692878851 |
- |
- |
- |
| 19 |
Hb_007423_020 |
0.2713417129 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 20 |
Hb_003906_170 |
0.2715002894 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |