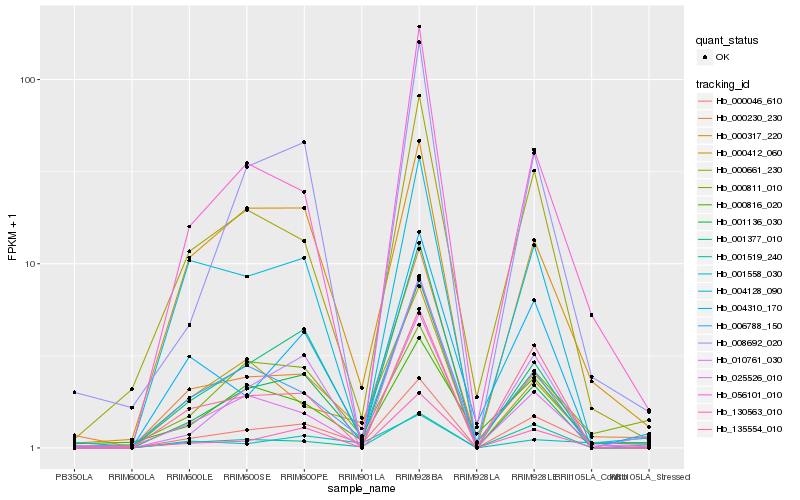
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_610 |
0.0 |
- |
- |
PREDICTED: putative cullin-like protein 2 isoform X3 [Populus euphratica] |
| 2 |
Hb_056101_010 |
0.1695730106 |
- |
- |
hypothetical protein POPTR_0391s00200g [Populus trichocarpa] |
| 3 |
Hb_025526_010 |
0.1736491116 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 4 |
Hb_010761_030 |
0.1767085616 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_000661_230 |
0.1774188532 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000230_230 |
0.1774900251 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 7 |
Hb_000317_220 |
0.1811566705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000811_010 |
0.1838230321 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF118 [Jatropha curcas] |
| 9 |
Hb_130563_010 |
0.1841196659 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_008692_020 |
0.1854746501 |
- |
- |
- |
| 11 |
Hb_001136_030 |
0.1862722836 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25-like [Jatropha curcas] |
| 12 |
Hb_000412_060 |
0.1872828881 |
- |
- |
hypothetical protein JCGZ_18814 [Jatropha curcas] |
| 13 |
Hb_006788_150 |
0.1887291118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001519_240 |
0.1929447669 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 15 |
Hb_001377_010 |
0.1930255344 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 16 |
Hb_004310_170 |
0.1933351295 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 17 |
Hb_004128_090 |
0.1952302097 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 18 |
Hb_000816_020 |
0.1962645047 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_135554_010 |
0.1967833988 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 20 |
Hb_001558_030 |
0.1969874929 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |