| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
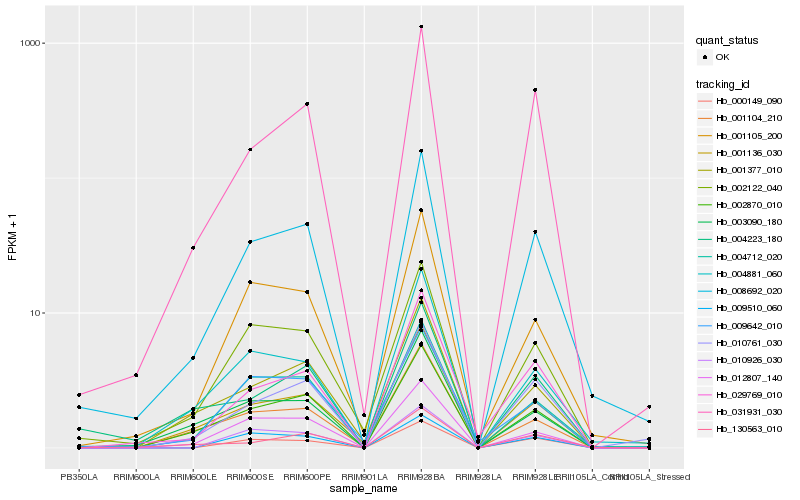
Hb_008692_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_004712_020 |
0.1039808797 |
- |
- |
hypothetical protein JCGZ_00782 [Jatropha curcas] |
| 3 |
Hb_010761_030 |
0.105867421 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_031931_030 |
0.1077544615 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 5 |
Hb_002122_040 |
0.1101875123 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 6 |
Hb_002870_010 |
0.1113246444 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 7 |
Hb_001105_200 |
0.1137616719 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_004881_060 |
0.1164429288 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 9 |
Hb_012807_140 |
0.1269977218 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |
| 10 |
Hb_130563_010 |
0.1270660382 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_029769_010 |
0.128481927 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 isoform X3 [Populus euphratica] |
| 12 |
Hb_000149_090 |
0.1312362859 |
- |
- |
- |
| 13 |
Hb_003090_180 |
0.1340693058 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 14 |
Hb_001136_030 |
0.1347585673 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25-like [Jatropha curcas] |
| 15 |
Hb_001377_010 |
0.1371418459 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 16 |
Hb_010926_030 |
0.1418285272 |
- |
- |
hypothetical protein CISIN_1g037967mg, partial [Citrus sinensis] |
| 17 |
Hb_009642_010 |
0.1435542197 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 18 |
Hb_001104_210 |
0.1437005587 |
- |
- |
hypothetical protein JCGZ_23620 [Jatropha curcas] |
| 19 |
Hb_009510_060 |
0.1473811089 |
- |
- |
- |
| 20 |
Hb_004223_180 |
0.1485723562 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |