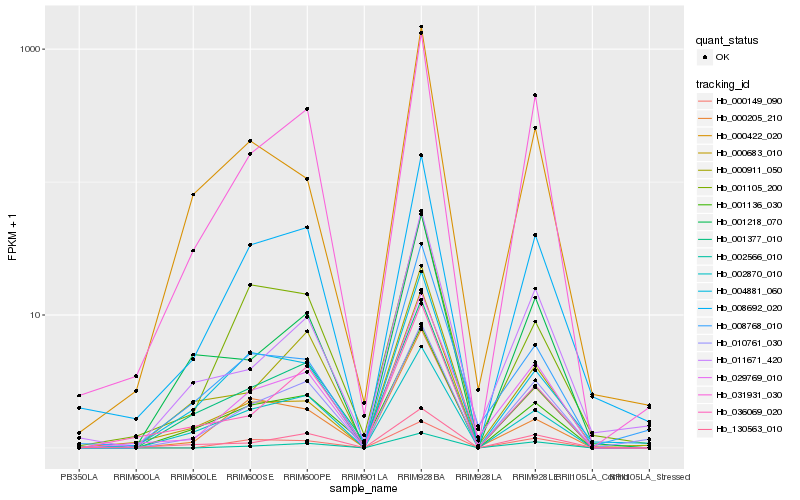
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029769_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 isoform X3 [Populus euphratica] |
| 2 |
Hb_031931_030 |
0.100524232 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 3 |
Hb_008692_020 |
0.128481927 |
- |
- |
- |
| 4 |
Hb_010761_030 |
0.1288845297 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_002566_010 |
0.1300549557 |
- |
- |
PREDICTED: uncharacterized protein LOC105643984 isoform X1 [Jatropha curcas] |
| 6 |
Hb_008768_010 |
0.1394443986 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000149_090 |
0.1418916523 |
- |
- |
- |
| 8 |
Hb_011671_420 |
0.1435581395 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 9 |
Hb_036069_020 |
0.1442610615 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 10 |
Hb_001136_030 |
0.1465969885 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25-like [Jatropha curcas] |
| 11 |
Hb_130563_010 |
0.1516530722 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001218_070 |
0.1516713268 |
- |
- |
PREDICTED: sugar transport protein 5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002870_010 |
0.1549481547 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 14 |
Hb_000683_010 |
0.1557153764 |
- |
- |
Interleukin-1 receptor-associated kinase, putative [Ricinus communis] |
| 15 |
Hb_000911_050 |
0.1617915394 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 21 [Jatropha curcas] |
| 16 |
Hb_001105_200 |
0.1619880902 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_004881_060 |
0.1627942313 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 18 |
Hb_000205_210 |
0.1632631757 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_001377_010 |
0.1635255889 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 20 |
Hb_000422_020 |
0.1700336014 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |