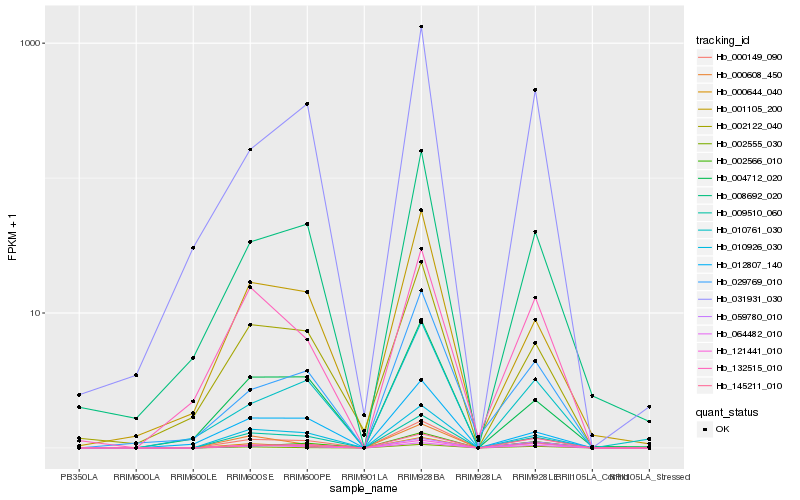
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000149_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_009510_060 |
0.0758461515 |
- |
- |
- |
| 3 |
Hb_010926_030 |
0.0777200418 |
- |
- |
hypothetical protein CISIN_1g037967mg, partial [Citrus sinensis] |
| 4 |
Hb_121441_010 |
0.1100066014 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 5 |
Hb_000644_040 |
0.1276323964 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_009157 [Vitis vinifera] |
| 6 |
Hb_002566_010 |
0.1289347821 |
- |
- |
PREDICTED: uncharacterized protein LOC105643984 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008692_020 |
0.1312362859 |
- |
- |
- |
| 8 |
Hb_064482_010 |
0.1349818545 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_002555_030 |
0.1368157147 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Malus domestica] |
| 10 |
Hb_031931_030 |
0.1385147272 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 11 |
Hb_004712_020 |
0.1414131962 |
- |
- |
hypothetical protein JCGZ_00782 [Jatropha curcas] |
| 12 |
Hb_029769_010 |
0.1418916523 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 isoform X3 [Populus euphratica] |
| 13 |
Hb_002122_040 |
0.1425789438 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_059780_010 |
0.1428087541 |
- |
- |
PREDICTED: uncharacterized protein LOC104885482, partial [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_010761_030 |
0.1454665874 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_001105_200 |
0.1493684886 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_012807_140 |
0.1552243059 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |
| 18 |
Hb_000608_450 |
0.1583756122 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 19 |
Hb_132515_010 |
0.1588793475 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 20 |
Hb_145211_010 |
0.1618127661 |
- |
- |
hypothetical protein M569_05648, partial [Genlisea aurea] |