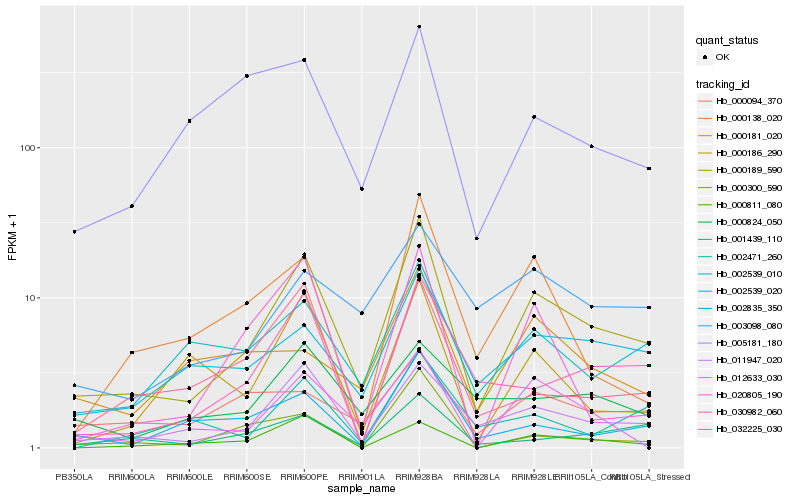
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_590 |
0.0 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 2 |
Hb_011947_020 |
0.1490327476 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 3 |
Hb_002539_020 |
0.1947187774 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 4 |
Hb_002539_010 |
0.1994935864 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 5 |
Hb_030982_060 |
0.1998162784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001439_110 |
0.2059722326 |
transcription factor |
TF Family: ERF |
PREDICTED: uncharacterized protein LOC105133481 [Populus euphratica] |
| 7 |
Hb_002835_350 |
0.2125553152 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 8 |
Hb_012633_030 |
0.2126461242 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |
| 9 |
Hb_000138_020 |
0.2239630507 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 10 |
Hb_000186_290 |
0.2258052612 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000094_370 |
0.2261597847 |
- |
- |
- |
| 12 |
Hb_000824_050 |
0.2269341869 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_032225_030 |
0.2273790805 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 14 |
Hb_000811_080 |
0.2279579344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_020805_190 |
0.2282674899 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_005181_180 |
0.2297234744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003098_080 |
0.2308397372 |
- |
- |
- |
| 18 |
Hb_002471_260 |
0.2328038341 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 19 |
Hb_000300_590 |
0.2337681926 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |
| 20 |
Hb_000181_020 |
0.2341130955 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |