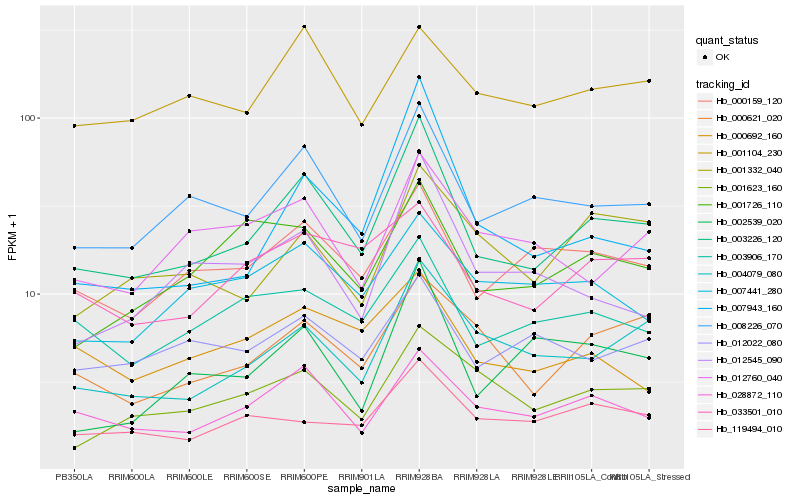
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003226_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008226_070 |
0.1053124848 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 3 |
Hb_028872_110 |
0.127049129 |
- |
- |
PREDICTED: uncharacterized protein LOC105644776 [Jatropha curcas] |
| 4 |
Hb_004079_080 |
0.1337133724 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 5 |
Hb_000159_120 |
0.1364699175 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 6 |
Hb_003906_170 |
0.1440197316 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_007943_160 |
0.148365271 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 8 |
Hb_000621_020 |
0.1500106811 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 9 |
Hb_012545_090 |
0.1512715915 |
- |
- |
hypothetical protein JCGZ_04320 [Jatropha curcas] |
| 10 |
Hb_001726_110 |
0.1520975158 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 19 [Jatropha curcas] |
| 11 |
Hb_002539_020 |
0.1536379995 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 12 |
Hb_033501_010 |
0.1541501115 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 13 |
Hb_001623_160 |
0.1552490993 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 14 |
Hb_119494_010 |
0.1559359778 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 15 |
Hb_012022_080 |
0.1588073737 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_001104_230 |
0.1596984716 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 17 |
Hb_012760_040 |
0.160798659 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 18 |
Hb_001332_040 |
0.1608297906 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 19 |
Hb_000692_160 |
0.160876503 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 20 |
Hb_007441_280 |
0.1615158779 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |