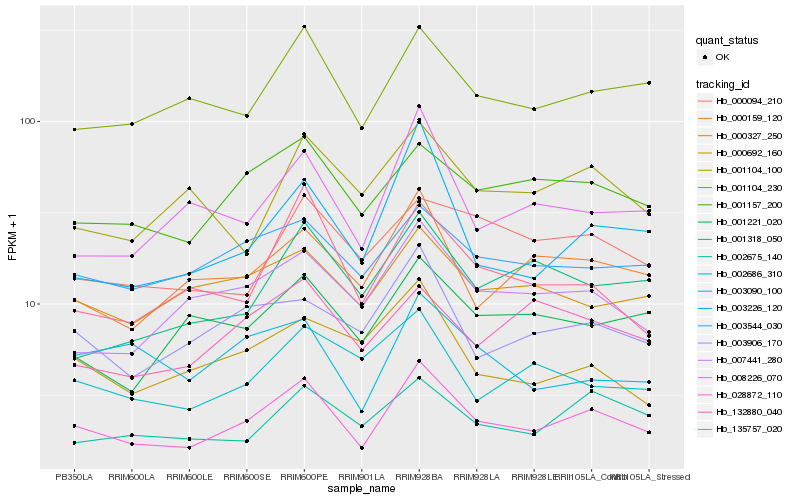
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028872_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644776 [Jatropha curcas] |
| 2 |
Hb_001104_230 |
0.1147829915 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 3 |
Hb_001157_200 |
0.1206286196 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 4 |
Hb_000159_120 |
0.1236139348 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 5 |
Hb_003906_170 |
0.1257569936 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_008226_070 |
0.1260384926 |
- |
- |
PREDICTED: expansin-A8 [Jatropha curcas] |
| 7 |
Hb_003226_120 |
0.127049129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003544_030 |
0.1282280139 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 9 |
Hb_007441_280 |
0.1283904675 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 10 |
Hb_002675_140 |
0.132906269 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 11 |
Hb_000327_250 |
0.1338010887 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 12 |
Hb_135757_020 |
0.1359994017 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 13 |
Hb_001221_020 |
0.1403171063 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 14 |
Hb_002686_310 |
0.1418922813 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 15 |
Hb_003090_100 |
0.1429385733 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 16 |
Hb_000692_160 |
0.1432602446 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 17 |
Hb_000094_210 |
0.1433688991 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 18 |
Hb_132880_040 |
0.1434274361 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001104_100 |
0.1451645087 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 20 |
Hb_001318_050 |
0.1453430568 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |