| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
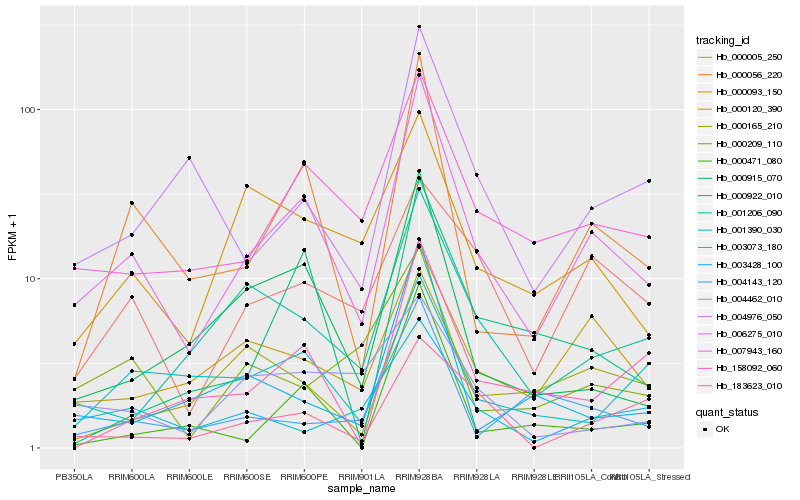
Hb_006275_010 |
0.0 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 2 |
Hb_000056_220 |
0.1560907741 |
- |
- |
PREDICTED: HVA22-like protein c [Jatropha curcas] |
| 3 |
Hb_007943_160 |
0.1725974218 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 4 |
Hb_000915_070 |
0.2049755144 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000120_390 |
0.211916611 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 6 |
Hb_001206_090 |
0.2205383934 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 7 |
Hb_004976_050 |
0.2218172939 |
- |
- |
- |
| 8 |
Hb_000093_150 |
0.2244338811 |
- |
- |
6-phosphogluconolactonase, putative [Ricinus communis] |
| 9 |
Hb_158092_060 |
0.2263284587 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000005_250 |
0.2285457626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000471_080 |
0.2303199906 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 12 |
Hb_000165_210 |
0.2331111236 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000922_010 |
0.2349835884 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 14 |
Hb_000209_110 |
0.2353608509 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 15 |
Hb_003428_100 |
0.2376286881 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 16 |
Hb_003073_180 |
0.2382819408 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 17 |
Hb_004143_120 |
0.2422766167 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 18 |
Hb_183623_010 |
0.2457461046 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 19 |
Hb_001390_030 |
0.2466229728 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 20 |
Hb_004462_010 |
0.2468371481 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |