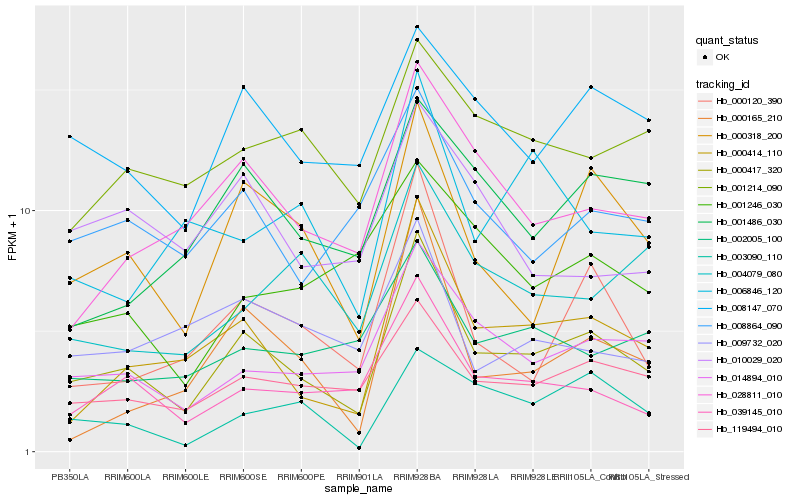
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_320 |
0.0 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 2 |
Hb_000414_110 |
0.1341289218 |
- |
- |
PREDICTED: MATE efflux family protein 5 isoform X1 [Populus euphratica] |
| 3 |
Hb_119494_010 |
0.1396772026 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 4 |
Hb_014894_010 |
0.1406692505 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 5 |
Hb_000318_200 |
0.1437130811 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 6 |
Hb_039145_010 |
0.1443437659 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 7 |
Hb_028811_010 |
0.1592403789 |
- |
- |
PREDICTED: uncharacterized protein LOC105650178 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008147_070 |
0.1629424468 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 9 |
Hb_000120_390 |
0.1673617268 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 10 |
Hb_008864_090 |
0.1679859019 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |
| 11 |
Hb_002005_100 |
0.1799790484 |
- |
- |
- |
| 12 |
Hb_001214_090 |
0.1835044048 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 13 |
Hb_003090_110 |
0.1851308519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001486_030 |
0.1853522232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_009732_020 |
0.1886383849 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 16 |
Hb_004079_080 |
0.1922905707 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 17 |
Hb_010029_020 |
0.1924576265 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 18 |
Hb_000165_210 |
0.1929245274 |
- |
- |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_006846_120 |
0.1953151426 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 20 |
Hb_001246_030 |
0.1956861358 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |