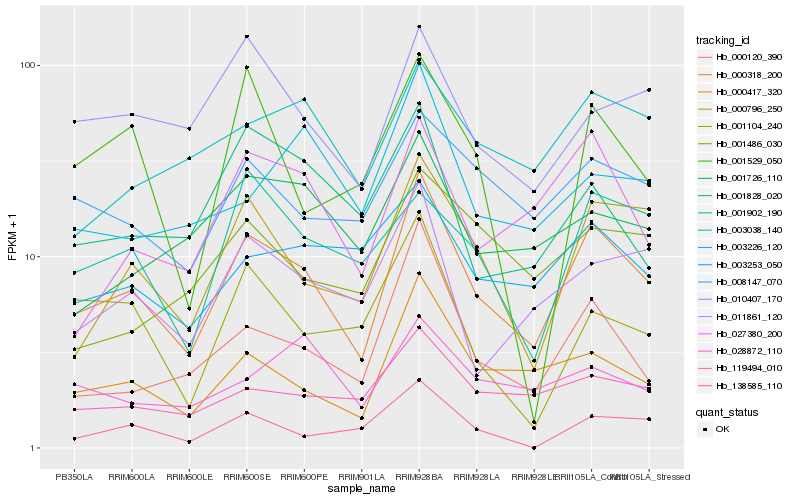
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_200 |
0.0 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 2 |
Hb_000417_320 |
0.1437130811 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 3 |
Hb_000796_250 |
0.1501582611 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 4 |
Hb_000120_390 |
0.1607534507 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 5 |
Hb_027380_200 |
0.1684125087 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 6 isoform X2 [Jatropha curcas] |
| 6 |
Hb_008147_070 |
0.168485981 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 7 |
Hb_119494_010 |
0.1689186759 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 8 |
Hb_011861_120 |
0.1701362298 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 9 |
Hb_010407_170 |
0.1776760356 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 10 |
Hb_003253_050 |
0.1808886003 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 3 [Jatropha curcas] |
| 11 |
Hb_003038_140 |
0.1814275532 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 12 |
Hb_001726_110 |
0.1855527174 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 19 [Jatropha curcas] |
| 13 |
Hb_001828_020 |
0.1888804947 |
- |
- |
hypothetical protein CISIN_1g010132mg [Citrus sinensis] |
| 14 |
Hb_138585_110 |
0.1901954388 |
- |
- |
- |
| 15 |
Hb_001902_190 |
0.1933095579 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 16 |
Hb_001486_030 |
0.1952158203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003226_120 |
0.1956958535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001104_240 |
0.1960174434 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_028872_110 |
0.1969850205 |
- |
- |
PREDICTED: uncharacterized protein LOC105644776 [Jatropha curcas] |
| 20 |
Hb_001529_050 |
0.2022591888 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |